Abstract
Fried syndrome, first described in 1972, is a rare X‐linked mental retardation that has been mapped by linkage to Xp22. Clinical characteristics include mental retardation, mild facial dysmorphism, calcifications of basal ganglia and hydrocephalus. A large four‐generation family in which the affected males have striking clinical features of Fried syndrome were investigated for linkage to X‐chromosome markers; the results showed that the gene for this condition lies within the interval DXS7109–DXS7593 in Xp22.2. In total, 60 candidate genes located in this region, including AP1S2, which was recently shown to be involved in mental retardation, were screened for mutations. A mutation in the third intron of AP1S2 was found in all affected male subjects in this large French family. The mutation resulted in skipping of exon 3, predicting a protein with three novel amino‐acids and with termination at codon 64. In addition, the first known large Scottish family affected by Fried syndrome was reinvestigated, and a new nonsense mutation, p.Gln66X, was found in exon 3. Using CT, both affected patients from the French family who were analysed had marked calcifications of the basal ganglia, as previously observed in the first Scottish family, suggesting that the presence of distinctive basal ganglia calcification is an essential parameter to recognise this syndromic disorder. It may be possible to use this feature to identify families with X‐linked mental retardation that should be screened for mutations in AP1S2.
Keywords: mental retardation, hydrocephalus, calcification of the basal ganglia, AP1S2, fried syndrome
The X‐linked condition hydrocephalus with aqueductal stenosis (HSAS) is the most common form of hereditary hydrocephalus (McKusick 30700), with an incidence of approximately 1 in 30 000, and accounts for around 2–15% of primary idiopathic hydrocephalus in male neonates.1 HSAS is associated with a wide variation in severity and a wide spectrum of clinical signs and symptoms both within and between families. Most cases with HSAS and those with MASA (mental retardation, aphasia, shuffling gait, and adducted thumbs) syndrome (McKusick 303350) are caused by heterogeneous mutations within the neural cell‐adhesion molecule L1 locus at Xq28.2 To date, around 34 families having known mutations in L1CAM have been described in the clinical literature, detailing a total of 129 people.3 However, several examples of families with HSAS suggest a genetic heterogeneity in this disease.4,5 Over 34 years ago, Fried described a Scottish family with X‐linked mental retardation (XLMR) associated with hydrocephalus or calcifications in the basal ganglia, or both.6,7 The family included six male patients with mental retardations in two generations. The degree of mental retardation varied from moderate to severe and there were two cases of hydrocephalus, one suspected to be the result of the stenosis of the aqueduct of Sylvius. All affected male patients had delayed motor development and difficulty in walking as adults. In 1996, linkage analysis mapped this X‐linked syndromic mental retardation locus to a region of 26.7 cM on Xp22 between markers DXS989 and KAL. Within this large interval, >70 characterised genes and expressed sequence tag (EST) clusters have been identified. In this paper, we report the description of a second family with clinical features reminiscent of those reported by Fried6,7 and the results of linkage studies refining the localisation of the gene in Xp22. We also report the results of sequencing of candidate genes located in this interval and the involvement of the AP1S2 gene recently found mutated in XLMR in the Fried syndrome.8
Methods
Clinical details
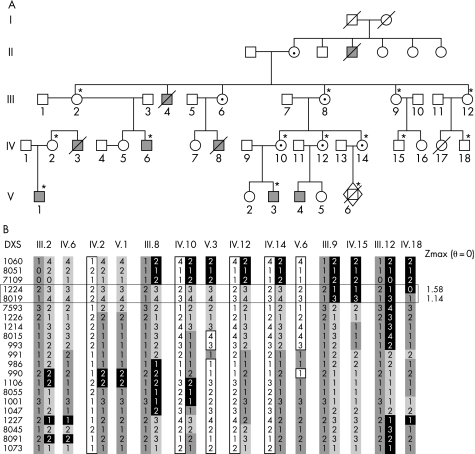
The pedigree is shown in fig 1A, with eight affected patients in four generations. The four living affected male patients live in different specialised institutions in France. Clinical findings were only available for five of the affected male patients.
Figure 1 (A) Pedigree of the family included in the linkage analysis. *Members for whom DNA was available and who were included in the linkage analysis. Filled squares, affected male patients; circles with dots, heterozygous female members. Female carrier status was assigned from family history (obligate carriers). (B) Haplotypes of the X‐chromosome markers and the recombined markers delimiting the potential genetic interval (horizontal rectangle). Maximum LOD scores are indicated on the right.
Patient VI
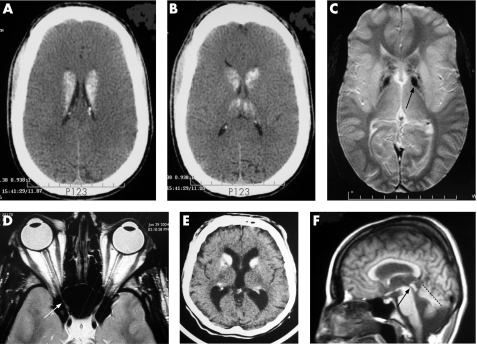
This patient was born in 1975 after a normal delivery (length 52 cm; weight 3.46 kg; head circumference 34.5 cm). He walked at 2.5 years of age. He has had severe behavioural problems from the age of 2 years and had severe hypotonia, auto‐aggressiveness and stereotypic hand movements. He has developed simple language (with single words) and attends a specialised institution. By the age of 20 years, he was capable of performing very simple tasks, but could walk only with help. At 29 years he suddenly lost visual acuity with irreversibly impaired vision, a result of major optic nerve atrophy due to osteosclerosis of the calvarium narrowing the optic canals (fog 2D). In addition, CT sand MRI scans showed marked bilateral calcification of the basal ganglia (mainly the caudate nucleus) and of the dendate nuclei of cerebellum (fig 2A, C).
Figure 2 Brain CT and MRI scans of affected patients: (A–D) VI and (E, F) IV6. (A, B) Axial CT scan: global severe thickening of the calvarium related to osteopetrosis, global caudate nucleus and patchy thalamus hyperdensity. (C) T2 gradient‐echo MRI: low heterogeneous signal in putamen and a signal in pallidum (black arrow). (D) Axial T2 scan in optic‐nerve plane: decreased calibre of optic nerve due to compression of the optic nerve within the optic canal (white arrow). (E) Axial CT scan: mild thickening of calvarium and hypredensity of the caudate nuclei. (F) Sagittal T2‐weighted MRI: mild ventricular dilatation without stenosis of acqueduc of Silvius (black arrow), and mild vermian hypoplasia (dotted line).
Patient IV6
This patient was born in 1966 without fetal distress after a normal pregnancy (birth weight 3650 g). He had neonatal hypotonia, and he walked unaided at 6 years old. He had profound mental retardation without any language. CT and MRI performed at 24 years of age revealed calcifications of the caudate nuclei, thick calvarium, mild ventricular dilatation without stenosis of acqueduc of Silvius, and mild vermian hypoplasia (fig 2E, F).
Patient III4
This patient was born in 1957, delivered at term by emergency caesarean section because of fetal distress. He was intubated at birth and was noted to have hypotonia as a neonate. He started to walk at the age of 14 months. He had seizures and a mild intellectual impairment (IQ of 71). He died at the age of 27 years.
Patients II4 and IV8
Both these patients had neonatal hypotonia and had severe disabilities. They died aged 1 and 2 years, respectively. Their death certificates stated congenital hydrocephalus with stenosis of the aqueduct of Sylvius.
DNA analysis
Genomic DNA was extracted from peripheral blood lymphocytes according to standard protocols. Blood samples were obtained after written consent from all tested individuals and/or their legal guardians. Microsatellite marker assay and linkage analysis were performed as described previously.5 Mutation screening in all coding exons and their corresponding flanking intron sequences of the different candidate genes was performed in affected patients.
DNA was amplified by PCR using 1× buffer II (Applied Biosystems, Foster City, California, USA), 1.5 mmol/l MgCl2, 200 μmol/l of each dNTP, 200 pmol of each gene‐specific primer (primer sequences are available upon request), 75 ng of DNA and 1.25 U of Taq polymerase (AmpliTaq Native; Applied Biosystems) on a PCR system (GeneAmp PCR system 9700 Gold; Applied Biosystems). Iinitial denaturation at 95°C for 7 min was followed by 40 cycles of denaturation for 15 s at 95°C, annealing for 15 s at the specific annealing temperature and elongation for 30 s at 72°C, and then a final extension at 72°C for 7 min. The fragments were directly sequenced or were analysed by denaturing high‐performance liquid chromatography (dHPLC). This method is a chromatographic mutation‐analysis technique that is based on temperature‐dependent separation of DNA containing mismatched base pairs from PCR‐amplified DNA fragments. When the procedures are carried out correctly (optimum PCR primer design, PCR protocol, separation gradient and oven temperature), almost 100% detection of single‐nucleotide variation and small deletion, insertion or duplication mutations can be achieved.9 PCR products showing an abnormal dHPLC profile were then directly sequenced on an automated sequencer (ABI 377; Applied Biosystems, Foster City, California, USA) using the dye‐terminator method. All variants were screened against a control panel of 160 X chromosomes to exclude polymorphisms.
AP1S2 mutation analysis was also performed by dHPLC in 300 patients with mental retartadation collected by the European XLMR Consortium.10
X‐chromosome inactivation status was ascertained using standard methods in female individuals who were carriers and in whom a mutation was identified. Briefly, aliquots of DNA were pre‐digested with the enzymes HpaII or CfoI. The triplet repeat at the HUMARA (human androgen receptor) locus was then amplified by PCR using fluorescent primers and analysed using an automated sequencer and genotyper software (ABI 3100; Applied Biosystems).11
mRNA analysis
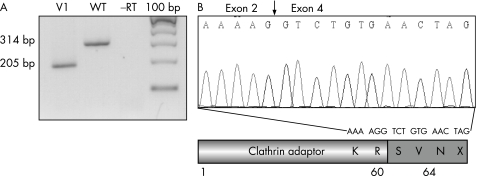
RNA isolation from lymphoblastoid cells from one of the affected patients (V1), and cDNA synthesis were performed according to standard protocols. We used primers designed to form cDNA products spanning exons 2–5 of AP1S2 gene (2F: 5′‐GAGTGGCGAGATCTGAAGAT‐3′ and 5R: 5′‐TGAAACCCCACGTAGTGTTC‐3′). Amplification was performed at an annealing temperature of 55°C to yield a 314 bp product. Reverse transcription PCR products were separated by electrophoresis through a 2% agarose gel, and products were sequenced on both strands using PCR primers with fluorescent dye terminators on an automated genetic analyser (ABI 3100; PE Applied Biosystems, Foster City, California, USA).
Results
Linkage analysis of the French family described in this study, using DNA markers evenly distributed in the X chromosome indicated linkage to Xp22, although only the marker DXS1224 had a LOD score of 1.58 with zero recombination (fig 2B). The interval included 60 candidate genes or expressed sequence tags located between DXS7109 and DXS7593 (about 10.4 Mb) (see http://genome.ucsc.edu and http://www.ensembl.org). Once linkage to Xp22 was established, candidate gene testing was initiated. We excluded from the molecular screening genes previously involved in other genetic diseases (such as TRAPPC2, OFD1, PIGA, RS1, PHEX, CDKL5, NHS, RPS6KA3, PHKA2, PDHA1). On the basis of their expression in fetal and/or adult brain, 22 genes (GPM6B, GLRA2, REPS2,EGFL6, GRPR, S100G, SYAP1, RBBP7, RAB9A, CXORF15, PPEF‐1, SH3KBP1, PDZK10, RAI2, EIF1AX, MAP3K15, FLJ14503, TMSB4X, MBTPS, YY2, SCML2 and CNKSR2) were tested. However, we found no causative mutations, but only several rare polymorphisms (data not shown). After a report was published describing nonsense mutations in the AP1S2 gene in patients with mild to severe mental retardation,8 this gene was also analysed as it falls within the linkage interval.
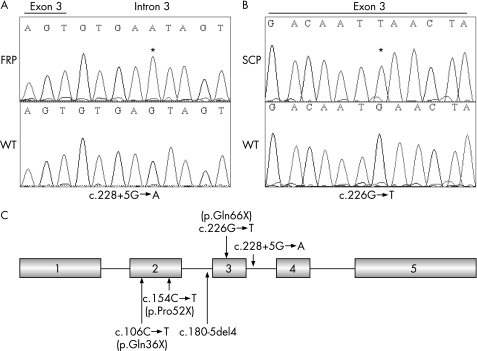
DNA analysis of male patient V1 was undertaken. Sequence analysis revealed a G→A transition at position +5 of the splice donor site in intron 3, c.288+5G→A (fig 3A). We analysed the segregation of the mutation in the family and confirmed that the mutation segregates with the phenotype. This result was consistent with the haplotype data that was derived from the linkage analysis. Furthermore, sequencing of 160 chromosomes from unaffected male individuals did not reveal this variation, indicating therefore that the c.288+5G→A transition is probably not a rare polymorphism. We studied X inactivation in DNA from blood cells of six unaffected female carriers (III.2, III.8, IV.2, IV.10, IV.12 and IV.14); the assay showed balanced X inactivation in all cases except one. Only the unaffected female member IV.2 had skewed X‐chromosome inactivation (5:95) (data not shown).
Figure 3 Mutational analysis in two families with Fried syndrome. Sequence chromatograms of exon 3 and flanking intron sequences of AP1S2 of a normal individual (wild type; wt) and an affected family member. Sequence variations are numbered starting from the first base of the ATG start codon, numbering based on the reference sequence NM_003916.3. (A) FRP (French patient), affected patient (V1) from the French Fried syndrome family; (B) SCP (Scottish patient), affected patient from the Scottish Fried syndrome family. Asterisk indicate the exon 2 to exon 4 junction. (C) Schematic representation of the exon structure of AP1S2, with positions of mutations previously found in families with XLMR and in the two families with Fried syndrome.
To assess the consequence of this potential mutation on AP1S2 mRNA splicing, RNA derived from lymphoblastoid cells of patient V1 and from a normal individual were used for RT‐PCR analysis. The normally spliced product is 314 bp in size; in patient V1, only a fragment of 205 bp was amplified (fig 4A). We found by direct sequencing of RT‐PCR products that only the amplified fragment from the mutated transcript was present (fig 4B), and the abnormal RT‐PCR fragment lacks the sequence corresponding to exon 3 (fig 4B). Skipping of exon 3 would then introduce a translational frameshift, resulting in three novel amino‐acids (SVN), with termination at codon 64 (fig 4B). Altogether, these results suggest that the mutation at the +5 position of the splice site completely abolishes normal splicing of exon 3 and results in truncated transcript with a premature stop codon.
Figure 4 (A) Reverse transcription (RT) PCR analysis using primers in exon 2 and 5 of the AP1S2 cDNA. Agarose‐gel electrophoresis of RT‐PCR products generated with lymphoblasts from patient (V1) and from a control subject (wild type; wt). (B) Sequence analysis of RT‐PCR products from the patient V1 and consequences of the splice mutation at the protein level.
In order to confirm the involvement of AP1S2 in Fried syndrome, we screened by direct sequencing the whole coding region of AP1S2 in the proband of the previously described Scottish family with Fried syndrome.7 Mutations searches identified one nonsense mutation located in exon 3 named p.Gln66X (c.226G→T) (fig 3B), which was not found in >160 normal X chromosomes. We analysed the segregation of the mutation in this family using DNA from three affected patients, one obligate carrier, and three non‐carrier female members (status inferred from analysis of haplotypes) and confirmed that the mutation segregates with the phenotype. This mutation is predicted to truncate the AP1S2 protein with termination at codon 66. To evaluate whether this gene was involved in non‐syndromic forms of mental retardation, we carried out AP1S2 mutation analysis by dHPLC on DNA samples of 300 affected patients collected by the European XLMR Consortium and found no pathogenic mutation (nor polymorphisms), suggesting that this gene is probably only involved in a syndromic phenotype. In 1 of the 160 normal X chromosomes, we found a C→T substitution at position 288; however, the resulting codon did not modify the corresponding amino acid (p.Ser96Ser).
Discussion
Recently, systematic screening of the coding exons and splice junctions of 737 annotated genes identified two nonsense mutations (p.Q36X, p.R52X) and one consensus splice‐site mutation (c.180‐5del4fsX64) in the AP1S2 gene on Xp22 in three unrelated families (fig 4C).8 All affected male patients have mental retardation, significant delay in walking, and marked hypotonia.
We report two unrelated (one French and one Scottish) families with common features of Fried syndrome and novel mutations in the AP1S2 gene. The phenotypes of the various members of the French family are variable but exhibit many similarities with the Scottish family. All affected male patients have mental retardation, poor speech and hypotonia. However, the degree of intellectual disability ranges from mild to profound. Interestingly, a CT scan of patients V1 and IV6 showed typical extensive calcification of the basal ganglia and dentate nuclei of the cerebellum, as described previously in the Scottish family. The CT scan also showed osteosclerosis of the calvarium, a rare skeletal condition characterised by increased bone density caused by aberrant osteoclast‐mediated bone resorption.12
The novel splice mutation c.288+5G→A identified in the French family produces the same truncated protein (stop codon at position 64) as the c.180‐5del4 previously found in family 63.8 However, in the Scottish family, all affected patients had mental retardation, microcephaly (head circumference 3–55), childhood hypotonia and no hydrocephalus. In this study, we found that the G→Α substitution at position +5 completely abolished normal splicing at least in lymphoblasts. The other mutation we identified was a novel nonsense mutation in exon 3, which can also produces a similar truncated protein (codon stop at position 66). To date, the two mutations in our families with Fried syndrome increase the number of unique mutations in the AP1S2 gene to five.
Using CT, we found marked calcifications of the basal ganglia in both analysed affected patients (patients V1 and IV6) from our French family, as previously observed in the Scottish family, the first described with Fried syndrome (patients III.26, III.35 and III.41), suggesting that the presence of the distinctive basal‐ganglia calcification is an essential parameter in diagnosis of this syndromic disorder. In the recent publication of Tarpey et a;,8 no radiological description was given for the affected patient in the three reported families with mutations in the AP1S2 gene. However, we can speculate that it is likely that several affected patients also had calcification of the basal ganglia. Because all affected male patients have mental retardation, marked hypotonia at age 6 months and understandable language, and a few patients (two in two unrelated families) have hydrocephalus, we can speculate that these three previously described families represent Fried syndrome. CT scans of these families would be useful to confirm this hypothesis. Calcification of basal ganglia and dentate nuclei of the cerebellum has also been observed in Fahr syndrome;13 however, in this heterogeneous condition, patients have movement disorders (such as rigid hypokinetic syndrome), dementia and other behavioural disturbances (such as mood disorders). Interestingly, the CT scan of patient V1 showed osteosclerosis of the calvarium. Osteosclerosis and cerebral calcification is also seen in carbonic anhydrase II deficiency or “marble brain” syndrome. This syndrome is also associated with mental retardation, short stature, and optic atrophy leading to blindness. There is constant metabolic acidosis, and a severe selective reduction of carbonic anhydrase in erythrocyte is often found.14 Interestingly, our patient V1 developed rapid and severe bilateral optic atrophy at the age of 29 years, due to osteosclerosis narrowing the optic canals within the small wings of the sphenoid bone. However, no feature associated with renal tubular acidosis is known in Fried syndrome.
The AP1S2 gene is composed of five exons and encodes a small protein of 157 amino acids, containing a large clathrin adaptor complex small‐chain domain and a small C‐terminal domain of unknown function.15 This protein is involved in formation of clathrin‐coated vesicles that form part of a well‐characterised system of eukaryotic membrane‐protein trafficking. The system has two major components: clathrin, which provides structural integrity, and adaptor protein complexes, which determine selection of the vesicle cargo and promote clathrin‐lattice formation onto the vesicle membrane. There are five adaptor protein complexes associated with clathrin, of which AP1 and AP2 are the best characterised. Both AP1 and AP2 adaptor complexes are composed of two large subunits, one medium subunit, and one small sigma subunit.15 AP1S2, which we have shown to be involved in Fried syndrome, encodes the sigma 2 subunit of the heterotetrameric AP1 adaptor complex. In yeast dual‐hybrid assays, it interacted with both gamma adaptin and gamma‐2 adaptin. These adaptor complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. The AP1 complex is preferentially associated with endosomes and the trans‐Golgi network, suggesting that a deficit in the function of AP1 should result in aberrant endocytic processing. It has been shown that the adaptor AP1 complex of the trans‐Golgi network clathrin interacts with microtubules and modulates the dynamics of the cytoskeleton, the stability and shape of coated organelles, and the loading of nascent AP1‐coated vesicles onto the appropriate microtubule tracks.16
Our study reinforces the fact that a defect in the trafficking of membrane proteins can impair brain development in the early stages by altering neural cell fate and/or disrupting regulation of cellular proliferation and apoptosis. We have shown that Fried syndrome, a severe neurological disorder characterised by severe mental retardation, spastic diplegia and hydrocephalus, is caused by mutations in the AP1S2 gene. CT scans of patients with Fried syndrome with mutations in AP1S2 show marked calcifications of the basal ganglia, suggesting that this parameter would be useful in identifying families with XLMR that should be screened for mutations in AP1S2.
Key points
Linkage analysis of a French family with X‐linked mental retardation associated with hydrocephalus and calcifications in basal ganglia mapped the locationto Xp22, between DXS7109 and DXS7593.
Evaluation of candidate genes in this interval showed mutation in the sigma 2 subunit of the adaptor protein 1 complex (AP1S2) gene.
Implication of this gene in the syndromic mental retardation family described by Fried was also demonstrated by identification of an additional AP1S2 mutation.
Patients with mental retardation with calcifications of the basal ganglia detected by CT should be screened for mutations in AP1S2.
Acknowledgements
The authors thank all the members of the EURO (European) X‐linked Mental Retardation (MRX) consortium. This work was supported by Institut National de la Santé et de la Recherche Médicale (ANR‐maladies rares ANR‐06‐MRAR‐003‐01).
Abbreviations
dHPLC - denaturing high‐performance liquid chromatography
EST - expressed sequence tag
HSAS - hydrocephalus with aqueductal stenosis
MASA - mental retardation, aphasia, shuffling gait, and adducted thumbs
RT - reverse transcription
XLMR - X‐linked mental retardation
Footnotes
Competing interests: None declared.
References
- 1.Schrander‐Stumpel C, Howeler C, Jones M, Sommer A, Stevens C, Tinschert S, Israel J, Fryns J P. Spectrum of X‐linked hydrocephalus (HSAS), MASA syndrome, aand complicated spastic paraplesia (SPG1)‐clinical review with 6 additional families. Am J Med Genet 199557107–116. [DOI] [PubMed] [Google Scholar]
- 2.Kenwrick S, Jouet M, Donnai D. X linked hydrocephalus and MASA syndrome. J Med Genet 19963359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamakasi M, Thompson P, Lemmon V. CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics 199728175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willems P J, Vits L, Raemaekers P, Beuten J, Coucke P, Holden J J A, Van Broeckhoven C, Warren S T, Sagi M, Robinson D, Dennis N, Friedman K J, Magnay D, Lyonnet S, White B N, Wittwer B H, Aylsworth A S, Reicke S. Further locaization of X‐linked hydrocephalus in the chromosomal region Xq28. Am J Hum Genet 199251307–315. [PMC free article] [PubMed] [Google Scholar]
- 5.Strain L, Wright A F, Bonthron D T. Fried syndrome is a distinct linked mental retardation syndrome mapping to Xp22. J Med Genet 199734535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried K. X‐linked mental retardation and/or hydrocephalus. Clin Genet 19723258–263. [PubMed] [Google Scholar]
- 7.Fried K, Sanger R. Possible linkage between Xg and the locus for a gene causing mental retardation with or without hydrocephalus. J Med Genet 19731017–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarpey P S, Stevens C, Teague J, Edkins S, O'Meara S, Avis T, Barthorpe S, Buck G, Butler A, Cole J, Dicks E, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, West S, Widaa S, Yates A, Catford R, Butler J, Mallya U, Moon J, Luo Y, Dorkins H, Thompson D, Easton D F, Wooster R, Bobrow M, Carpenter N, Simensen R J, Schwartz C E, Stevenson R E, Turner G, Partington M, Gecz J, Stratton M R, Futreal P A, Raymond F L. Mutations in the gene encoding the sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X‐linked mental retardation. Am J Hum Genet 2006791119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donohoe E. Denaturing high‐performance liquid chromatography using the WAVE DNA fragment analysis system. Methods Mol Med 2005108173–187. [DOI] [PubMed] [Google Scholar]
- 10.De Brouwer A P, Yntema H G, Kleefstra T, Lugtenberg D, Oudakker A R, de Vries B B, Van Bokhoven H, Van Esch H, Frints S G, Froyen G, Fryns J P, Raynaud M, Moizard M P, Ronce N, Bensalem A, Moraine C, Poirier K, Castelnau L, Saillour Y, Bienvenu T, Beldjord C, des Portes V, Chelly J, Turner G, Fullston T, Gecz J, Kuss A W, Tzschach A, Jensen L R, Lenzner S, Kalscheuer V M, Ropers H H, Hamel B C. Mutations frequencies of X‐linked mental retardation genes in families from the Euro MRX consortium. Hum Mutat 200728207–208. [DOI] [PubMed] [Google Scholar]
- 11.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen‐receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992511229–1239. [PMC free article] [PubMed] [Google Scholar]
- 12.Kocher M S, Kasser J R. Osteopetrosis. Am J Orthop 200332222–228. [PubMed] [Google Scholar]
- 13.Modrego P J, Mojonero J, Serrano M, Fayed N. Fahr's syndrome presenting with pure and progressive presenile dementia. Neurol Sci 200526367–369. [DOI] [PubMed] [Google Scholar]
- 14.Shah G N, Bonapace G, Hu P Y, Strisciuglio P, Sly W S. Carbonic anhydrase II deficiency syndrome (osteopetrosis with renal tubular acidosis and brain calcification): novel mutations in CA2 identified by direct sequencing expand the opportunity for genotype‐phenotype correlation. Hum Mutat 200424272. [DOI] [PubMed] [Google Scholar]
- 15.Takatsu H, Futatsumori M, Yoshino K, Yoshida Y, Shin H W, Nakayama K. Similar subunit interactions contribute to assembly of clathrin adaptor complexes and COPI complex: analysis using yeast three‐hybrid system. Biochem Biophys Res Commun 20012841083–1089. [DOI] [PubMed] [Google Scholar]
- 16.Orzech E, Livshits L, Leyt J, Okhrimenko H, Reich V, Cohen S, Weiss A, Melamed‐Book N, Lebendiker M, Altschuler Y, Aroeti B. Interactions between adaptor protein‐1 of the clathrin coat and microtubules via type 1a microtubule‐associated proteins. J Biol Chem 200127631340–31348. [DOI] [PubMed] [Google Scholar]