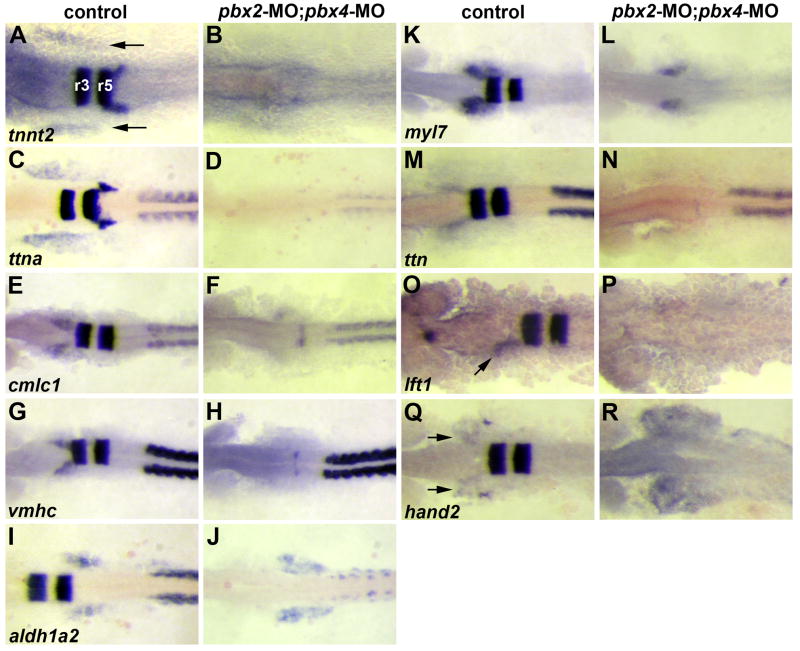
Fig. 2.
Validation of microarray-identified Pbx-dependent heart genes. (A–R) RNA in situ expression of genes from Table 1 in (A,C,E,G,I,K,M,O,Q) wild-type control or (B,D,F,H,J,L,N,P,R) pbx2-MO; pbx4-MO embryos. (A–D) are at 10 somite stage (10s). (E–N, Q–R) are at 18s. (O–P) are at 20s to achieve more robust expression of lft1 in controls. krox-20, labeled in hindbrain rhombomeres r3 and r5 in (A), is included in all in situs to control for loss of Pbx. The wild-type heart primordium expression domains of tnnt2, ttna, cmlc1, vmhc, myl7, lft1 and hand2 are just anterior-lateral to r3 krox-20 expression (arrows in A,O,Q). n≥10 for each marker in control or pbx2-MO; pbx4-MO. Embryos are shown in dorsal view, anterior towards the left.