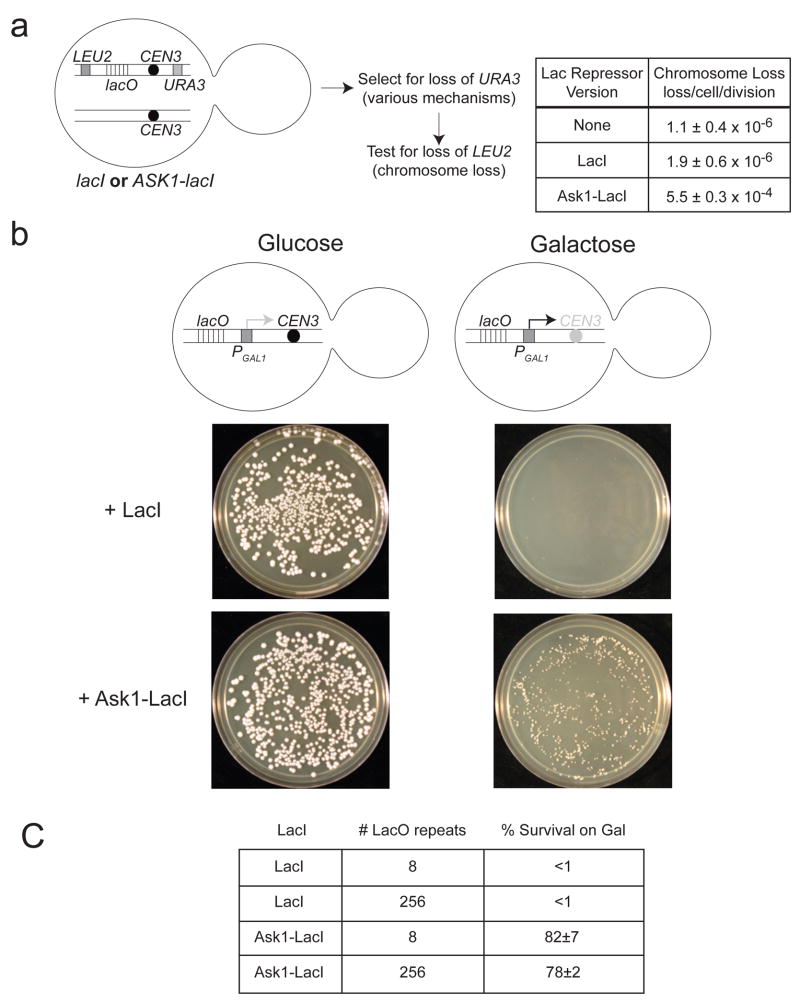
Figure 2. The synthetic kinetochore can replace a natural kinetochore.
A) The presence of a synthetic kinetochore interferes with normal chromosome segregation. The cartoon shows the relevant features of the genotype of a diploid strain carrying one normal copy of chromosome III and one that carried both the natural and synthetic kinetochore. We measured the loss frequency of this chromosome in cells where the synthetic kinetochore is active (due to expression of the Ask1-LacI fusion) and those where it is not (cells without LacI and cells expressing unfused LacI). We first looked for loss of URA3 and then looked for those that had also lost LEU2 to determine that the entire chromosome was lost. The quoted range represents the 95% confidence interval. B) The synthetic kinetochore can substitute for a natural kinetochore. Cartoon of a conditional centromere. Galactose-induced transcription from the GAL1 promoter inactivates CEN3, rendering the natural kinetochore functional on glucose but inactive on galactose. Only cells expressing the Ask1-LacI fusion form colonies on galactose plates. Pictures were taken after 3 days of growth on glucose plates and 5 days of growth on galactose plates at 30°C. C) 8 or 256 repeats of the LacO array give similar results. Plating experiments were performed on strains with 8 or 256 repeats of the LacO array. Approximately 500 cells were plated to YPGlu or YPGal. The percent of cells that survived on YPGal compared to YPGlu was calculated and the average taken from 3 independent experiments where 4 plates were counted per experiment ± s.d.