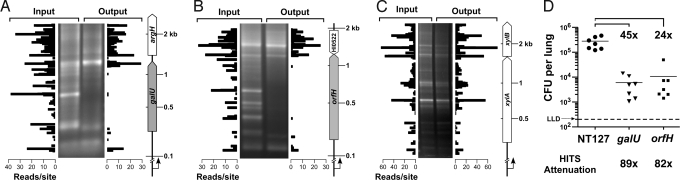
Fig. 3.
Comparison of HITS analysis, genetic footprinting, and single-strain infections. Genetic footprinting of input and output libraries for (A) galU, encoding UDP-glucose pyrophosphorylase, (B) orfH, encoding heptosyltransferase III, and (C) xylA, encoding xylose isomerase, are shown in the gel images. PCR analysis was conducted using the transposon-specific primer marout and chromosomal primers galU_F, orfH_F, or xylA_F that anneal 278 bp, 202 bp, and 279 bp upstream of the respective genes. In the plots, HITS data correspond to regions analyzed by footprinting. (D) H. influenzae NTHi wild-type (NT127) and deletion mutants of galU and orfH recovered from lungs of C57BL/6 mice (7 mice per strain) 24 h after intranasal inoculation with each strain. Bars represent the mean cfu per lung. Comparisons between wild-type and mutants were statistically significant via one-way ANOVA with Tukey's multiple comparison test (P < 0.001). LLD, lower limit of detection. Fold differences in mean cfu recovered for NT127 wild-type strain vs. the galU or orfH mutants in individual mutant infections (brackets) are compared with HITS results (below the chart). HITS survival indices were 0.011 for galU and 0.012 for orfH, corresponding to in vivo attenuations (calculated as the reciprocal of the s.i.) of 89-fold and 82-fold, respectively. In A–C, genome coordinates of transposon insertion sites detected via HITS analysis were reoriented with respect to the chromosomal primer positions used in footprinting. The y axis was modeled to the migration of the molecular weight (MW) standard of footprinting gels using nonlinear regression, and the x axis represents the number of sequencing reads mapped to insertion sites. The scale of the MW standards on the right of each panel applies to both the genetic footprints and the HITS analysis plots. White, nonessential genes; gray, genes required for growth or survival in the lung.