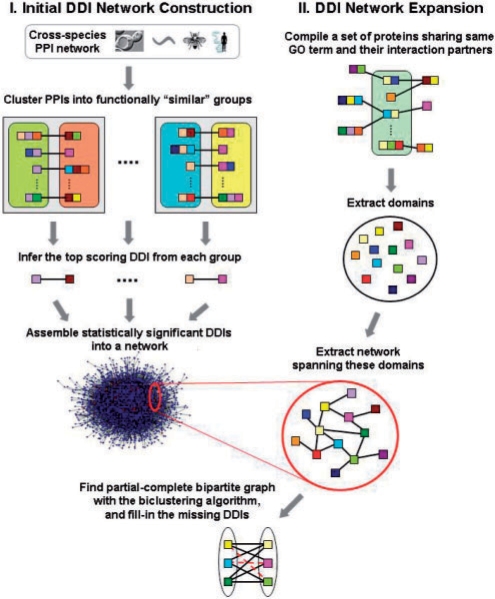
Fig. 1.
A schematic of the K-GIDDI method to infer DDIs. Colored squares denote protein domains, and proteins are denoted by single or concatenated domains. PPIs and DDIs are denoted by edges/lines connecting proteins or domains, respectively. Colored rounded-rectangles contain proteins sharing the same GO ‘molecular function’ term, and gray squares containing a pair of rounded-rectangles denote groups containing functionally similar PPIs. A representative DDI pattern is derived from each group. Those inferred DDIs that pass the statistical significance thresholds are then assembled to generate the initial DDI network, which is then expanded via a knowledge-guided search to include DDIs that may not be identified otherwise by methods that rely only on PPI data. The network expansion procedure, guided by GO information, identifies proteins sharing the same GO terms and their interaction partners, extracts the network spanning the domains within these proteins, and uses a biclustering algorithm to identify partial-complete bipartite sub-network and fills-in missing edges, denoted as red broken lines, to make them complete bipartite sub-networks.