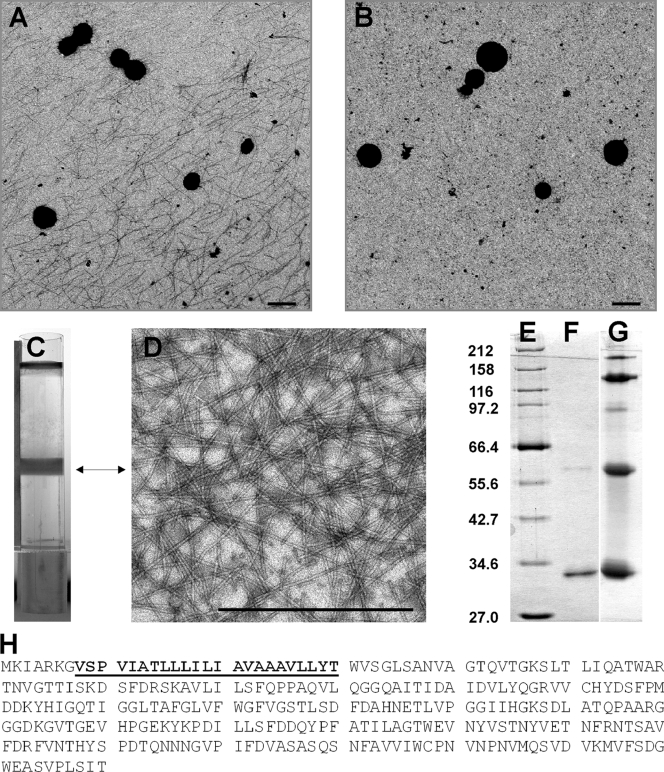
FIG. 1.
Ultrastructural and biochemical analyses of Ignicoccus hospitalis fibers. (A) A 5-μl aliquot of a fermentor culture, grown to stationary phase, was applied onto a carbon-coated grid, negatively stained with 2% uranyl acetate, and analyzed by TEM. The sample was collected using a wide-bore pipette. (B) An aliquot of the same culture as that in panel A was analyzed by the TEM procedure as described for panel A. The sample was collected with a syringe and needle. (C) CsCl gradient (volume, 10 ml) of I. hospitalis fibers concentrated by PEG precipitation from 10 liters of supernatant of a 16,000 × g centrifugation (used to harvest cells from the fully grown fermentor culture). (D) TEM analysis of the CsCl gradient-purified fibers, prepared and negatively stained as for panel A. (E to G) SDS-PAGE analyses of CsCl gradient-purified fibers (12.5% gel, silver stained). (E) Broad-range protein marker (NEB). Sizes in kDa are indicated to the left. (F) Heat-denatured fibers (0.5 μg treated for 15 min at 100°C). (G) Partially heat-denatured fibers (5 μg treated for 3 min at 100°C). (H) Amino acid sequence of the fiber protein. The sequence given in boldface and underlined was obtained by N-terminal protein sequencing and used to identify Igni_0670 as the structural gene coding for the I. hospitalis fibers (NCBI reference sequence NC_009776.1). The size bars in panels A, B, and D are 1 μm each.