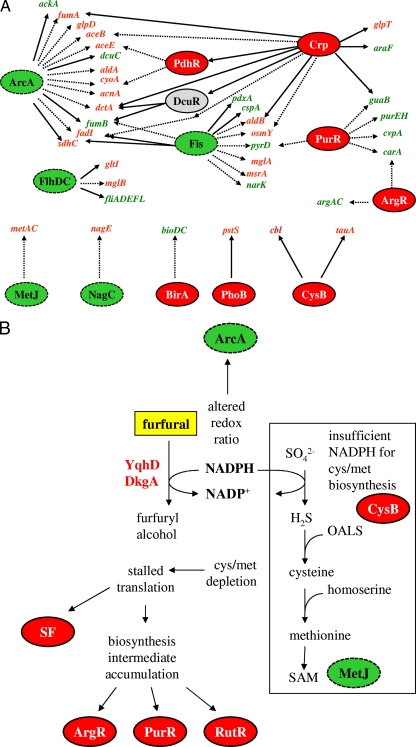
FIG. 1.
Transcriptional and regulatory changes in LY180 following challenge with 0.5 g liter−1 furfural. Regulatory genes that were significantly perturbed were identified by NCA with a P value cutoff of 0.05 relative to a null distribution. Regulators with increased activity are shown in red with a solid border, regulators with decreased activity are shown in green with a dashed border. Regulators that showed a mixed activity are shown in gray. Table 3 gives a more comprehensive listing of all perturbed regulators. (A) Partial regulator gene response map. Representative genes that were perturbed more than twofold are shown, with red (green) indicating genes with increased (decreased) expression. Solid lines indicate activation by the connected regulator, and dashed lines indicate repression. Because the direction of perturbation for DcuR is unclear, this regulator is shown in gray. (B) Model illustrating the mechanism of growth inhibition by furfural. The addition of furfural induces two NADPH-dependent oxidoreductases (YqhD and DkgA) that compete with essential biosynthetic reactions for NADPH. Assimilation of sulfate requires 4 NADPH molecules per cysteine and appears to be the most vulnerable of these biosynthetic reactions, as evidenced by perturbation of CysB and MetJ and by the direct benefit of supplementation with alternative sulfur sources. Secondary consequences include depletion of sulfur amino acids and a cascade of events from stalled translation and accumulation of many non-sulfur building block intermediates to a more general stress response, as evidenced by perturbation of the stringent factor (SF) and biosynthesis regulators ArgR, PurR, and RutR. OALS, O-acetyl-l-serine; SAM, S-adenosylmethionine.