Abstract
TRIM5α is a tripartite motif (TRIM) protein that consists of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains. The TRIM5αrh protein from rhesus monkeys recognizes the human immunodeficiency virus type 1 (HIV-1) capsid as it enters the host cell and blocks virus infection prior to reverse transcription. HIV-1-restricting ability can be eliminated by disruption of the B-box 2 domain. Changes in the TRIM5αrh B-box 2 domain have been associated with alterations in TRIM5αrh turnover, the formation of cytoplasmic bodies and higher-order oligomerization. We present here the nuclear magnetic resonance structure of the TRIM5 B-box 2 domain and identify an unusual hydrophobic patch (cluster 1) on the domain surface. Alteration of cluster 1 or the flanking arginine 121 resulted in various degrees of inactivation of HIV-1 restriction, in some cases depending on compensatory changes in other nearby charged residues. For this panel of TRIM5αrh B-box 2 mutants, inhibition of HIV-1 infection was strongly correlated with higher-order self-association and binding affinity for capsid complexes but not with TRIM5αrh half-life or the formation of cytoplasmic bodies. Thus, promoting cooperative TRIM5αrh interactions with the HIV-1 capsid represents a major mechanism whereby the B-box 2 domain potentiates HIV-1 restriction.
Mammalian proteins that can dominantly block retroviral infection by interfering with the early phase of viral replication have been identified (1a, 47, 50). One member of the tripartite motif (TRIM) family of proteins, TRIM5α, mediates species-specific restriction of particular retroviruses from diverse retrovirus subfamilies. For example, TRIM5α from rhesus macaques (TRIM5αrh) blocks infection of human immunodeficiency virus type 1 (HIV-1) soon after viral entry but prior to reverse transcription (22, 50). Although human TRIM5α (TRIM5αhu) only weakly inhibits HIV-1 infection, this protein potently inhibits infection by N-tropic murine leukemia virus (N-MLV) (16, 39, 42, 47, 57). Some species of Old World and New World monkeys encode TRIMCyp instead of TRIM5α; TRIMCyp is a fusion of TRIM5 and cyclophilin A and restricts some retroviruses, including HIV-1 (4, 16, 38, 39, 42, 47, 55-57).
TRIM5α is composed of RING, B-box 2, and coiled-coil domains typical of TRIM proteins and a carboxy-terminal B30.2(SPRY) domain. The RING domain is not absolutely required for, but contributes to the potency of, retroviral restriction; the RING domain plays an important role in the rapid degradation of TRIM5α (11). Some changes in the B-box 2 domain exert dramatic effects on retrovirus-restricting ability (10, 19, 40). The coiled-coil domain allows oligomerization of TRIM5α (20, 36), which recently has been shown to be a dimer (16, 39, 42, 47, 57). Dimerization contributes to the avidity of TRIM5α for the HIV-1 capsid (51). The B30.2(SPRY) domain, which mediates capsid recognition, dictates the viral specificity of restriction (46, 52, 58). Sequence divergence within variable regions of the B30.2(SPRY) domain accounts for species-specific differences in the potency of restriction (46, 52, 58).
Structural and functional similarities exist between the RING and B-box domains of TRIM proteins. Both domains bind two zinc atoms tetrahedrally in a cross-brace conformation (3, 17, 34, 35). Furthermore, both domains have been shown to be involved in protein-protein interactions. In non-TRIM proteins, RING domains play the role of molecular scaffolds, allowing formation of supramolecular complexes by self-association, which in some cases improves enzymatic activity (2, 23, 24, 43). Interestingly, RING-RING interactions have been reported to be functionally important for genomic stability, as in the heterodimer formed by the RING domain of BRCA1 with the RING domain of BARD-1 (5).
The B-box domain is found in extremely diverse proteins; in TRIM proteins, the B-box domain is present in one or two copies (44, 54). B-box domains have been implicated in protein-protein interactions (32, 53) and have been shown to play a role in protein localization, growth control, differentiation, and transcriptional regulation (54). The B-box domain usually adopts a ββα conformation reminiscent of the RING domain. Some TRIM proteins, such as MID1 (TRIM18), contain two B-box domains in tandem. Structural studies demonstrated that the tandem B-boxes of MID1 pack against each other (53). The contact interface between the two B-boxes is made by the short β-strands of each B-box domain. Mutations affecting either one of these B-box domains occur in patients with Opitz G/BBB syndrome, a severe developmental disorder (48). Recent structural studies of the B-box 2 domain of MuRF1 (TRIM63) identified a dimer interface that consists of a hydrophobic surface and polar residues (46, 52, 58). These findings suggest that B-box-B-box interactions may have functional significance.
Using models based on the B-box 1 or B-box 2 structures of other proteins, mutagenesis of putative surface-exposed residues of the TRIM5αrh B-box 2 domain has been conducted (10, 19, 40). These studies have revealed several functions dependent upon the intact TRIM5αrh B-box 2 domain. First, the wild-type TRIM5α proteins of humans and rhesus monkeys are turned over rapidly, with a half-life of less than 1 h (11). Alteration of specific basic residues in the TRIM5αrh B-box 2 domain results in much more stable proteins; because some of these mutants retain antiretroviral activity, rapid turnover of TRIM5α appears not to be essential to retrovirus-restricting ability (10).
Second, a change in the B-box 2 domain of TRIM5α can affect the higher-order self-association of TRIM5α dimers (32). This self-association contributes to the ability of TRIM5α to bind the retroviral capsid, perhaps by increasing avidity (32). The contribution of the B-box 2 domain to antiretroviral activity is particularly important when the interactions of the B30.2(SPRY) domain with the viral capsid are weak (31, 32).
Like many TRIM proteins (44), TRIM5αrh forms aggregates called cytoplasmic bodies when overexpressed (11). A third phenotype associated with TRIM5α B-box 2 changes is a decrease in the propensity of the protein to coalesce into cytoplasmic bodies; this phenotype is particularly striking, because many TRIM5α B-box 2 mutants are expressed at higher steady-state levels than the wild-type protein (10). Although cytoplasmic bodies per se are not necessary for TRIM5α-mediated retroviral restriction (46, 52, 58), the relationship between higher-order TRIM5α self-association, which contributes to capsid binding (32), and the tendency to form cytoplasmic aggregates is not clear.
Lastly, some TRIM5α B-box 2 mutants apparently lose the ability to block retrovirus infection out of proportion to any measured reductions in capsid binding (10, 32). This observation hints that the B-box 2 domain may mediate functions beyond higher-order self-association and capsid binding. Presumably, such functions could contribute mechanistically to the process whereby TRIM5α disrupts the early events in retrovirus infection following the recognition of the capsid after it enters the cytosol. Studies following the fate of the HIV-1 capsid in the cytosol of infected cells have correlated restriction of infection with a decrease in the amount of cytosolic particulate capsids (51). Similar changes have been observed in the amount of cytosolic particulate capsids during infection of HIV-1 and N-MLV in the presence of the restriction factors TRIMCyp and TRIM5αhu, respectively (10, 41). In contrast to the wild-type TRIM5αhu protein, a TRIM5αhu B-box 2 mutant defective for N-MLV restriction did not cause a reduction in the amount of particulate capsid in the cytosol (46, 52, 58). Thus, B-box 2 functions may contribute to the accelerated uncoating of the retroviral capsid associated with TRIM5α restriction.
We present here a nuclear magnetic resonance (NMR) structure of the TRIM5 B-box 2 domain and demonstrate a weak tendency of the purified B-box 2 domains to self-associate. Alteration of the unusually hydrophobic surface or flanking charged amino acid residues of the TRIM5αrh B-box 2 domain revealed structures important for rapid turnover, higher-order self-association, cytoplasmic body formation, capsid binding, and retrovirus restriction. To understand which B-box 2 functions contribute to retroviral restriction, the relationships among these TRIM5α properties were investigated. Both higher-order self-association and binding to the viral capsid strongly correlated with inhibition of HIV-1 infection, implying that the B-box 2 domain potentiates retroviral restriction by promoting cooperative TRIM5α interactions with the capsid.
MATERIALS AND METHODS
Sample preparation of the TRIM5 B-box 2 domain.
The DNA fragment encoding the B-box 2 of human TRIM5α (amino acid residues S86 to T131 [Swiss-Prot Q9C035]) was amplified via PCR from the Research Association for Biotechnology Clone FLJ14687 and was cloned into the plasmid vector pCR2.1 (Invitrogen, Carlsbad, CA) as a fusion with an N-terminal His tag and a tobacco etch virus protease cleavage site. The 13C- and 15N-labeled protein was synthesized by a cell-free protein expression system (25). Purification was performed by a standard procedure (29). For structure determination, a single 1.13 mM uniformly 13C- and 15N-labeled sample was prepared in 20 mM Tris-HCl buffer (pH 7.0), 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.02% NaN3, 0.05 mM ZnCl2, and 1 mM iminodiacetic acid, with the addition of D2O to 10% (vol/vol). The engineered protein sample used for the NMR measurements was comprised of 46 amino acid residues from human TRIM5 spanning the B-box 2 domain (residues 86 to 131 of TRIM5αhu); the first seven and last six residues of the protein consist of tags. The residues of this engineered protein are referred to as 79 to 137, so that the numbers of the B-box 2 residues correspond to those of human TRIM5.
NMR spectroscopy, structure determination, and analysis.
All of the NMR spectra were recorded at 20°C on Varian INOVA 600, 800, and 900 spectrometers equipped with pulsed-field gradient triple-resonance probes. Sequence-specific resonance assignments were made using the standard triple-resonance techniques. The backbone assignment was achieved by the combined analysis of HNCO, (HCA)CO(CA)NH, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra. The side chain resonances were identified by the combinational use of HBHANH, HBHA(CO)NH, (H)CC(CO)NH, (H)CCH-TOCSY, HCCH-TOCSY, and two-dimensional 1H-15N HSQC and 1H-13C HSQC spectra. Nuclear Overhauser effect (NOE) data for structure determination were extracted from three-dimensional 15N- and 13C-edited NOESY spectra, recorded with mixing times of 75 and 65 ms, respectively.
The NMRpipe software package (7) and the program KUJIRA (26), created on the basis of NMRView (21), was used for optimal visualization and spectral analysis. Automated NOE cross-peak assignments (18) and structure calculations with the torsion angle dynamics were performed using the software package CYANA2.0.17 (15).
Dihedral angle restraints were derived using the program TALOS (6). A total of 100 conformers were calculated independently. The 20 conformers with the lowest final CYANA target function values were finally selected. The structures were validated by using PROCHECK-NMR (28). The program MOLMOL (27) was used to analyze the resulting 20 conformers and to prepare drawings of the structures, unless noted otherwise in the legends.
The selected 20 conformers have been deposited in the Protein Data Bank (PDB entry 2YRG).
Dynamic light-scattering experiment.
The data were measured at 4°C by using a DynaPro light scattering system (Protein Solutions, Lakewood, NJ) and a microsampler, with a protein concentration of 2.2 mg/ml in 20 mM Tris-HCl buffer (pH 7.0) containing 100 mM NaCl and 1 mM iminodiacetic acid. Dynamics Version 5.0 software (Protein Solutions) was used in the data collection and analysis.
Analytical ultracentrifugation.
Sedimentation equilibrium experiments were performed on a Beckman Optima XL-I centrifuge, in an eight-position An-50Ti rotor with equilibrium six-channel centerpieces and quartz windows. Experiments were carried out at 4°C with the B-box 2 domain protein in solution at concentrations of 1.2, 0.8, 0.4, and 0.2 mg/ml and at rotor speeds of 20,000, 22,000, and 24,000 rpm for the dimeric molecular weight range and 30,000, 31,000, and 32,000 rpm for the monomeric molecular weight range. The data were corrected after 12, 14, and 16 h at each speed. Protein concentration profiles were monitored by the absorbance at 280 nm. To generate the baseline, the samples were then completely sedimented at 40,000 rpm for 6 h at 4°C. For the calculation of the molecular weight, corrected datasets were analyzed in the self-association model with a partial specific volume of 0.705 ml/g and a solvent density of 1.003 g/ml, using the Beckman XL-A/XL-I data analysis software (version 6.03).
Creation of cells stably expressing TRIM5α variants.
Retroviral vectors encoding wild-type or mutant rhesus monkey TRIM5αrh proteins were created using the pLPCX vector. The TRIM5αrh proteins contained an influenza hemagglutinin (HA) epitope tag at the C terminus or FLAG epitope tags at the N terminus. Recombinant viruses were produced in 293T cells by cotransfecting the pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells (59). Cf2Th canine thymocytes were transduced and selected in 5 μg of puromycin (Sigma)/ml.
Infection with viruses expressing GFP.
Recombinant HIV-1 and N-MLV expressing green fluorescent protein (GFP) were prepared as described previously (12, 50). All recombinant viruses were pseudotyped with the vesicular stomatitis virus G glycoprotein. For infections, 3 × 104 Cf2Th cells seeded in 24-well plates were incubated at 37°C with virus for 24 h. Cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScan (Becton Dickinson). HIV-1 and N-MLV viral stocks were titrated by serial dilution on Cf2Th cells to determine the concentration of infectious viruses.
Protein analysis.
Cellular proteins were extracted with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 2 mg of aprotinin/ml, 2 mg of leupeptin/ml, 1 mg of pepstatin A/ml, 100 mg of phenylmethylsulfonyl fluoride/ml). The cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE; 10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia Biotech). Detection of protein by Western blotting utilized monoclonal antibodies directed against the hemagglutinin (HA) epitope tags (Roche) and FLAG epitope tags (Sigma) and monoclonal antibodies to β-actin (Sigma). All monoclonal antibodies were directly conjugated to horseradish peroxidase. Proteins were detected by enhanced chemiluminescence (NEN Life Sciences Products) using the FluorChem FC2 detection system (Alpha Innotech). Signals were acquired as an image file (TIFF) and quantified by using QuantityOne software (Bio-Rad Laboratories).
TRIM5α dimerization.
Approximately 107 293T cells transfected with 5 μg of plasmids expressing TRIM5αrh variants were lysed in NP-40 lysis buffer (0.5% NP-40 in phosphate-buffered saline [PBS]) and cross-linked with different concentrations (0to 10 mM) of ethylene glycolbis-succinimidylsuccinate (EGS) for 30 min as previously described (10). The cross-linking reaction was stopped by adding 10 μl of 1 M Tris-HCl. After cross-linking, the samples were resuspended in 2× sample buffer, followed by incubation at 37°C for 30 min. Samples were analyzed by SDS-PAGE and Western blotting with an anti-HA antibody (Roche).
Higher-order self-association of TRIM5α.
Human 293T cells were independently transfected with plasmids encoding FLAG-tagged and HA-tagged mutant and wild-type TRIM5αrh proteins. After 48 h, the cells expressing each TRIM5αrh variant were lysed in 1 ml of whole-cell extract buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% IGEPAL-10% glycerol, 1 mM DTT, protease inhibitor cocktail [Roche]). Lysates were centrifuged at 14,000 rpm for 1 h at 4°C. Post-spin lysates were then precleared using protein A-agarose (Sigma) for 1 h at 4°C; a small aliquot of each of these lysates was stored as an INPUT sample. Precleared lysates containing the differently tagged proteins were mixed in a 1/1 ratio and incubated with anti-FLAG-agarose beads (Sigma) for 2 h at 4°C to precipitate the FLAG-tagged proteins. Beads containing the immunoprecipitate were washed four times in whole-cell extract buffer. Subsequently, immune complexes were eluted using 200 μg of FLAG tripeptide/ml in whole-cell extract buffer. The eluted samples were separated by SDS-PAGE and analyzed by Western blotting using anti-HA or anti-FLAG antibodies conjugated to horseradish peroxidase.
TRIM5α turnover.
The turnover rates of wild-type and mutant TRIM5α proteins were estimated by pulse-chase experiments. Cf2Th cell lines stably expressing TRIM5 proteins were metabolically labeled for 30 min with 0.1 mCi of [35S]methionine-cysteine (35S protein-labeling mix; Perkin-Elmer)/ml in Dulbecco modified Eagle medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. The cells were then chased for different time intervals in Dulbecco modified Eagle medium containing an excess of unlabeled methionine and cysteine. Cells were subsequently lysed in a modified RIPA buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, 0.25% SDS). Samples were subsequently immunoprecipitated with anti-HA antibody-Sepharose beads for 16 h at 4°C. After two washes with modified RIPA buffer, samples were separated by SDS-PAGE (Invitrogen) and analyzed by autoradiography and PhosphorImager (Molecular Dynamics).
Localization of wild-type and mutant TRIM5α proteins.
The localization of TRIM5αrh and variants was investigated as previously described (8). Briefly, cells were grown overnight on 12-mm-diameter coverslips and fixed in 3.9% paraformaldehyde (Sigma) in PBS (Cellgro) for 30 min. Cells were washed in PBS, incubated in 0.1 M glycine (Sigma) for 10 min, washed in PBS, and permeabilized with 0.05% saponin (Sigma) for 30 min. Samples were blocked with 10% donkey serum (Dako, Carpinteria, CA) for 30 min and incubated for 1 h with antibodies. The anti-HA fluorescein isothiocyanate-conjugated 3F10 antibody (Roche) was used to stain HA-tagged proteins. Subsequently, samples were mounted for fluorescence microscopy by using the ProLong Antifade kit (Molecular Probes, Eugene, OR). Images were obtained with a Bio-Rad Radiance 2000 laser scanning confocal microscope with Nikon 60× N.A.1.4 optics.
HIV-1 CA-NC expression and purification.
The HIV-1 CA-NC protein was expressed, purified, and assembled as previously described (13, 14). The pET11a expression vector (Novagen) expressing the CA-NC protein of HIV-1 was used to transform BL-21(DE3) E. coli. CA-NC expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an optical density of 0.6 at 600 nm. After 4 h of induction, the cells were harvested and resuspended in 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, 10 mM 2-mercaptoethanol, and protease inhibitors (Roche). Lysis was performed by sonication, and debris was pelleted for 30 min at 35,000 × g. Nucleic acids were stripped from the solution by using 0.11 equivalents of 2 M (NH4)2SO4 and the same volume of 10% polyethylenimine. Nucleic acids were removed by stirring and centrifugation at 29,500 × g for 15 min. The protein was recovered by addition of 0.35 equivalents of saturated (NH4)2SO4. The protein was centrifuged at 9,820 × g for 15 min and resuspended in 100 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol. The CA-NC protein was dialyzed against 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol, and stored at −80°C.
In vitro assembly of CA-NC complexes.
HIV-1 CA-NC particles were assembled in vitro by diluting the CA-NC protein to a concentration of 0.3 mM in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 2 mg of DNA oligo-(TG)50/ml. The mixture was incubated at 4°C overnight and centrifuged at 8,600 × g for 5 min. The pellet was resuspended in assembly buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl) at a final protein concentration of 0.15 mM (13, 14) and stored at 4°C until needed.
Binding of TRIM5αrh variants to HIV-1 capsid complexes.
293T cells were transfected with plasmids expressing wild-type or mutant TRIM5αrh proteins. At 48 h after transfection, cell lysates were prepared as follows: previously washed cells were resuspended in hypotonic lysis buffer (10 mM Tris [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). The cell suspension was frozen, thawed, and incubated on ice for 10 min. Afterward, the lysate was centrifuged at full speed in a refrigerated Eppendorf microcentrifuge (∼14,000 × g) for 5 min. The supernatant was supplemented with 1/10 volume of 10× PBS and then used in the binding assay. In some cases, samples containing the TRIM5αrh variants were diluted with extracts prepared in parallel from untransfected cells. To test binding, 5 μl of CA-NC particles assembled in vitro were incubated with 200 μl of cell lysate at room temperature for 1 h. A fraction of this mixture was stored (input). The mixture was spun through a 70% sucrose cushion (70% sucrose, 1× PBS, and 0.5 mM DTT) at 100,000 × g in an SW55 rotor (Beckman) for 1 h at 4°C. After centrifugation, the supernatant was carefully removed and the pellet was resuspended in 1× SDS-PAGE loading buffer (pellet). The level of TRIM5αrh proteins was determined by Western blotting with an anti-HA antibody, as described above. The level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting with an anti-p24 CA antibody.
Quantitative real-time PCR.
Cf2Th cells expressing wild-type and mutant TRIM5αrh proteins were challenged with HIV-1 at a multiplicity of infection of 0.2. Viruses were pretreated with DNase to prevent contamination from carryover plasmid DNA. An infection using a virus that was heat-inactivated (60°C for 30 min) was performed as a control for carryover plasmid DNA in the PCR. After 6 h, the cells were lysed and DNA was extracted by using a Qiagen blood tissue DNA extraction kit. PCRs were prepared by using the QuantiTect probe PCR kit. Each sample contained 100 ng of total cellular DNA. PCR was carried out using two primers that amplify a 263-bp fragment of GFP: GFP-fwd, 5′-GAC GTA AAC GGC CAC AAG-3′; GFP-rev, 5′-GGT CTT GTA GTT GCC GTC GT-3′; and GFP-Probe, 5′-56-FAM-CCT ACG GCA AGC TGA CCC TGA-36-TAMRA-3′. A calibration curve was prepared using an HIV-1-GFP plasmid.
RESULTS
Solution structure of the TRIM5 B-box 2 domain.
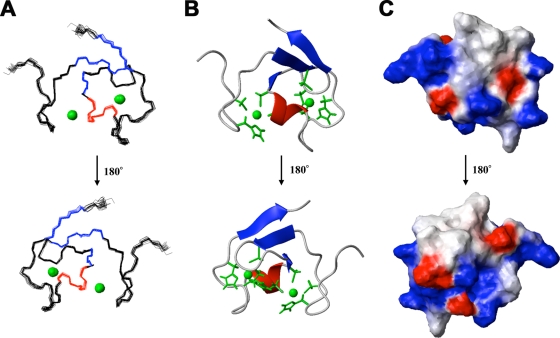
A fragment (residues 86 to 131) of human TRIM5 that encompasses the B-box 2 domain was expressed and labeled in a cell-free system. The solution structure of the monomeric protein was determined by using multidimensional NMR spectroscopy (PDB entry 2YRG). The 1H-15N and 1H-13C HSQC spectra were broad for the size of the protein, but all of the resonances were assigned except for the amide proton of Q121. The solution structures were well defined from residues 93 to 131 (Fig. 1). The root mean square deviations (RMSDs) from the mean structure were 0.25 ± 0.07 Å for backbone atoms and 0.83 ± 0.10 Å for heavy atoms in the defined region of the domain. The structural statistics are summarized in Table 1 . Like most B-box domains (34, 35), the B-box 2 domain from TRIM5 adopts a ββα RING-like fold, which is maintained by two zinc ions and CHCDC2H2-type zinc coordination. There is also an additional very short β-strand just before the helix.
FIG. 1.
Solution structure of the human TRIM5 B-box 2 domain. (A) Superposition of 20 NMR structures showing the α-helix in red, the β-strands in blue, and the Zn2+ in green. (B) Ribbon diagram of the NMR structure shown in panel A from the same perspective. The Zn2+ coordinating residues (CHCDC2H2) are also shown in green. (C) Electrostatic mapping of the B-box 2 surface, highlighting the positions of the positive (blue), negative (red), or neutral (white) charges.
TABLE 1.
Summary of conformational constraints and statistics of the final 20 structures of the B-box 2 domain from TRIM5α
| Parameter | Value |
|---|---|
| NOE upper distance restraints | |
| Short range (|i − j| ≤ 1) | 461 |
| Medium range (1 < |i − j| < 5) | 185 |
| Long range (|i − j| ≥ 5) | 317 |
| Total | 963 |
| Dihedral angle restraints (φ and ψ) | 106 |
| CYANA target function value (Å2) | 0.09 |
| Ramachandran plot (%) | |
| Residues in most favored regions | 79.5 |
| Residues in additional allowed regions | 20.3 |
| Residues in generously allowed regions | 0.3 |
| Residues in disallowed regions | 0.0 |
| RMSD ± SD from the averaged coordinatesa (Å) | |
| Backbone atoms | 0.25 ± 0.07 |
| Heavy atoms | 0.83 ± 0.10 |
That is, for residues 93 to 131.
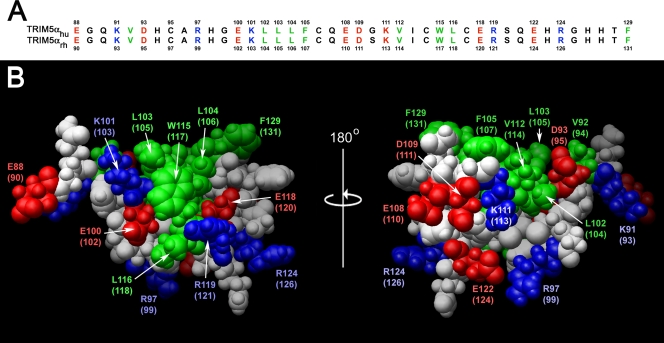
A noteworthy feature of the TRIM5α B-box 2 domain is that the majority of hydrophobic residues are solvent exposed and clustered, forming two hydrophobic patches on the domain surface. These hydrophobic surface patches are conserved in the B-box 2 domains of TRIM5α proteins from different species; for example, the human and rhesus monkey B-box 2 domains differ in only one amino acid residue, which does not reside within either hydrophobic patch (Fig. 2). One of the hydrophobic patches (cluster 1) is located on the outer surface of the helix and the first β-strand and is formed in human TRIM5α by residues L103, L104, W115, and L116. The cluster 1 patch is surrounded by the negatively charged residues E118 and E100 and positively charged residues R119 and K101. The second hydrophobic patch (cluster 2) is formed in human TRIM5α by L102, V112, F105, and F129.
FIG. 2.
B-box 2 domains of human and rhesus monkey TRIM5. (A) The human and rhesus monkey TRIM5 B-box 2 domain sequences are aligned and numbered. Residues with surface-accessible side chains are colored as follows: green, hydrophobic; red, acidic; and blue, basic. (B) A Corey-Pauling-Koltun model of the human and rhesus monkey TRIM5 B-box 2 domain is shown. The residue numbers correspond to those of human TRIM5, with the residue numbers of rhesus monkey TRIM5 in parentheses. The orientations of the left and right figures are the same as those of the upper and lower figures, respectively, in Fig. 1; the right image represents a rotation of the left image 180° around the vertical axis. Acidic and basic residues are shown in red and blue, respectively. Hydrophobic residues are shown in green. Cluster 1 is evident in the left figure, and cluster 2 is evident in the right figure.
Concentration dependence of NMR signals.
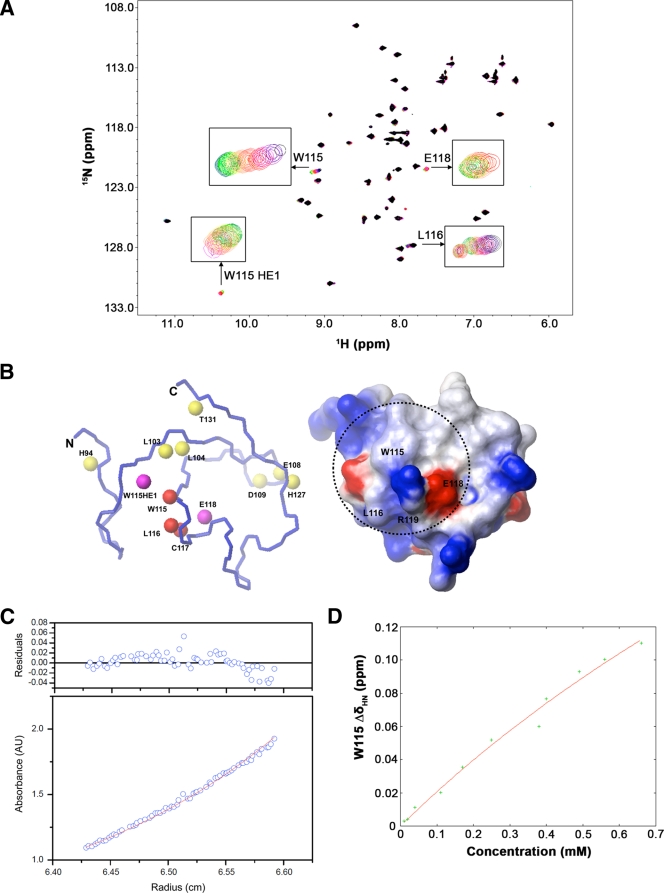
The residues on the human TRIM5 B-box 2 protein exhibited broadened signals in the 1H-15N HSQC spectrum; this contrasts with previous observations with other B-box 2 domains, which usually give sharp signals because of their compact size and stable fold (34). To investigate the basis of the observed broadening, the concentration dependence of the 1H-1D and 1H-15N HSQC spectra was measured (Fig. 3A). As the protein concentration decreased, the peaks of the amide protons from K101, L116, and W115 and side chain CH protons from L116 were shifted; the broadened peaks of the amide protons from L104 and E118, and the imino proton from the W115 indole ring, were sharpened or appeared. These results indicate that the B-box 2 domains associated via the cluster 1 region in the millimolar range of concentration and dissociated at lower concentrations (Fig. 3B). We also observed additional chemical shift perturbations (CSPs) in the very low concentration range <100 μM) involving the very narrow region near E108 and H127; this phenomenon may result from the zinc-mediated B-box 2 association at H127 (see below).
FIG. 3.
HSQC perturbation analysis in concentration-dependent experiments. (A) Eleven 1H-15N HSQC spectra were measured at protein concentrations of 660, 560, 490, 400, 300, 250, 170, 110, 40, 20, and 10 μM and superimposed, with colors ranging from purple (highest concentration) to green (lowest concentration). Signal appearances were observed for the backbone amide proton of E118 and imino proton of the W115 indole ring at the lower concentrations. Chemical shift changes and sharpening were observed for other signals. (B) Mapping of perturbed residues on the B-box 2 structure. In the figure on the left, the size of the CSP is indicated by color. The red color means more than 0.03 ppm of CSP were observed, and yellow indicates more than 0.01 ppm of CSP. The magenta color indicates the appearance of the signal at a lower concentration. Most of the implicated residues are proximal to cluster 1 on one face of the B-box 2 domain (dotted circle on the electrostatic surface on the right). (C) Equilibrium analytical ultracentrifugation of the TRIM5 B-box 2 domain. The radial distribution of the absorbance in the centrifuge cell at equilibrium at 22,000 rpm, with a protein concentration of 1.2 mg/ml (∼180 μM), is shown in the lower panel. The red line through the data represents the fit to a single species with a molecular weight corresponding to 10,502; the theoretical molecular weight for the single polypeptide chain is 6,682. The upper panel shows the residuals for the fit. (D) The concentration dependence of chemical shift perturbation for the W115 backbone amide proton in 1H-15N HSQC spectra. Total chemical shift change was calculated by using the relation: Δδtot = [(δHN)2 + (δN/6.5)2]1/2 (1). Curve fitting was carried out by using the program gnuplot. The calculated equilibrium dissociation constant KD was 9.33 ± 5.40 mM (see the text).
Size and affinity of the associated state of the B-box 2 domain.
To estimate the oligomeric state of the associated B-box 2 domain in solution, we performed a high-speed sedimentation equilibrium study by analytical ultracentrifugation (Fig. 3C). The radial distributions at 20,000, 22,000, and 24,000 rpm with a B-box 2 domain concentration of 1.2 mg/ml (∼180 μM) fitted to a molecular mass of 1.05 × 104 Da, which corresponds to 1.57 molecules of the B-box 2 domain. This implies that the B-box 2 domain exists between monomeric and dimeric forms in solution at this concentration. In addition, dynamic light scattering experiments revealed that the B-box 2 domain in solution had a monomodal profile with a narrow peak corresponding to particles with an average hydrodynamic diameter of 3.3 nm and an estimated molecular mass of 11 kDa (data not shown). Although it is difficult to calculate an exact molecular mass by dynamic light scattering, this result is consistent with the TRIM5 B-box 2 domain existing in solution at an average size between those of monomers and dimers. The partial dimer content of TRIM5 B-box 2 solutions may reflect a zinc-mediated self-association of the B-box 2 domain, because we detected a chemical shift change specifically in H127 in the low protein concentration range (<100 μM); in contrast, we did not observe any additional chemical shift changes in other residues in the concentration range from 10 to 660 μM (data not shown). We also found that reducing the amount of the weak chelator iminodiacetic acid in the buffer resulted in a much higher average molecular mass in the sedimentation equilibrium measurement and reduced solubility of the B-box 2 protein (data not shown). These results support the interpretation that most of the dimer estimated from the sedimentation equilibrium data resulted from zinc-mediated association. Collectively, the results suggest two types of B-box association, a zinc-mediated association and a direct association involving the region from W115 to E118. The zinc-mediated association appears to exhibit a higher affinity but affects the HSQC spectra less. The direct association has a lower affinity but causes major concentration-dependent shifts of NMR peaks that reflected the dynamic equilibrium between zinc-mediated associates.
Since the zinc-mediated association did not cause CSPs except for H127, the CSP data in the other areas of the domain could be fitted independently. The equilibrium dissociation constant (KD) was calculated to be 9.33 ± 5.40 mM using gnuplot in the dimer model by the analysis of chemical shift changes of the W115 amide proton in the concentration-dependent two-dimensional 1H-15N HSQC spectra (Fig. 3D). The fitting error was very large because of the low affinity and limited solubility of the protein. We estimated that only ca. 10% of the protein contributed to dimerization even at the highest concentration (0.66 mM). This explains the small observed chemical shift changes, even though the indole ring of W115 is apparently included in the binding interface. The small chemical shift changes of the backbone amide protons also suggest that the association does not involve a change of the protein fold and is stabilized by inter-side chain interactions.
Mutagenic analysis of the TRIM5αrh B-box 2 surface.
To investigate the functional implications of the TRIM5 B-box structure, we modified specific amino acid residues of TRIM5αrh. We chose the TRIM5αrh protein because it potently restricts HIV-1 infection (50), allowing quantitation of phenotypes over a wide dynamic range. Moreover, most of the assays to evaluate TRIM5α phenotypes have been developed using the rhesus monkey protein as a prototype (11, 32, 36, 50, 51). Because the TRIM5αrh and TRIM5αhu B-box 2 primary amino acid sequences differ by only one residue (see the alignment in Fig. 2A), structure-function relationships are expected to be similar between these two domains. Amino acid residues predicted to be surface accessible on the TRIM5αrh B-box 2 domain, including those predicted to reside in clusters 1 and 2, were altered (Fig. 2B). Particular attention was paid to the charged residues surrounding cluster 1, with various combinations of charge substitutions being introduced into this region. The wild-type and mutant TRIM5αrh proteins were expressed stably in Cf2Th canine thymocytes, which do not express a TRIM5 ortholog (45). All of the TRIM5αrh mutants studied were expressed at least as well as the wild-type TRIM5αrh protein (see Fig. S1 in the supplemental material). The steady-state levels of expression of several of the B-box 2 mutants were greater than that of wild-type TRIM5αrh, which has a half-life of ∼48 min (see Fig. S2 in the supplemental material). Indeed, prolonged half-lives, relative to that of the wild-type TRIM5αrh, were associated with many of the TRIM5α mutants (Table 2). This result is consistent with previous studies that implicated the TRIM5α B-box 2 domain in the regulation of TRIM5α turnover (10, 11). All of the TRIM5α mutants tested formed dimers detected by EGS cross-linking (see Fig. S3 in the supplemental material and Table 2). This result is consistent with previous studies indicating that the TRIM5α B-box 2 domain can be deleted without disrupting dimerization (10).
TABLE 2.
Phenotypes of TRIM5α B-box 2 mutants
| TRIM5αrh variant | Restrictiona
|
Mean reverse transcription (%) ± SDb,h | Dimerizationc,h | Higher-order association (%)d,h
|
Mean binding to HIV-1 CA-NC complexes ± SDe,h | Localizationf | Half-life (min)g | ||
|---|---|---|---|---|---|---|---|---|---|
| HIV-1 | N-MLV | Association with wild type | Self-association | ||||||
| Wild type | ++++ | ++ | 0 | Yes | 100 | 100 | 1 ± 0 | Wild-type | 48 |
| E120D | ++++ | + | 0 | ND | 100 | ND | 1.26 ± 0.05 | L reticular | 80 |
| L104E | ++++ | + | 0 | Yes | ND | ND | 1.09 ± 0.29 | L reticular | >1,000 |
| K93A | ++++ | + | 0 | Yes | ND | ND | 1.23 ± 0.04 | Wild-type | 60 |
| Q92A | ++++ | + | 0 | Yes | ND | ND | 1.14 ± 0.12 | Wild-type | 65 |
| F131E | ++++ | + | ND | ND | ND | ND | 0.84 ± 0.06 | L reticular | 60 |
| Q123E | ++++ | - | ND | ND | ND | 73 | 1.00 ± 0.30 | Wild-type | 70 |
| R121K | ++++ | - | 0 | ND | 7 | 81 | 1.34 ± 0.07 | Wild-type | 42 |
| R99E | ++++ | - | 0 | Yes | ND | 85 | 0.95 ± 0.23 | L reticular | 240 |
| K103E | ++++ | - | 0 | ND | ND | ND | 0.90 ± 0.08 | L reticular | >1,000 |
| L105E | +++ | - | ND | ND | ND | ND | 1.04 ± 0.16 | Reticular | >1,000 |
| E120R/R121E | +++ | - | 56.5 ± 10 | Yes | ND | 15 | 0.76 ± 0.15 | Reticular | >1,000 |
| E120D/R121K | +++ | - | 25 ± 11 | ND | 3 | ND | 1.12 ± 0.24 | Reticular | 400 |
| E120K/R121K | +++ | - | ND | ND | 0 | 70 | 0.71 ± 0.12 | L reticular | 110 |
| L132E | +++ | - | 63.5 ± 7 | ND | 0 | 0 | 0.84 ± 0.13 | L reticular | 120 |
| E102R | +++ | - | ND | ND | ND | ND | 0.97 ± 0.23 | M reticular | 400 |
| E102R/R121E | +++ | - | 81.5 ± 26 | Yes | 0 | 28 | 0.86 ± 0.04 | M reticular | 600 |
| E120R | ++ | - | ND | ND | 6 | ND | 0.45 ± 0.12 | M reticular | ND |
| E120R/R121K | ++ | - | ND | ND | 0 | 80* | 0.45 ± 0.06 | L reticular | 240 |
| E120K | ++ | - | 5 ± 5 | ND | 0 | 14 | 0.84 ± 0.10 | L reticular | 300 |
| E102R/E120R/R121E | ++ | - | ND | ND | 0 | ND | ND | L reticular | 400 |
| E120D/R121D | ++ | - | ND | Yes | 0 | ND | 0.85 ± 0.06 | L reticular | 360 |
| E120R/R121D | ++ | - | 19 ± 11 | ND | 0 | ND | 0.72 ± 0.10 | Reticular | 400 |
| E120K/R121E | + | - | ND | Yes | 0 | 5 | 0.73 ± 0.07 | L reticular | >1,000 |
| E120K/R121D | + | - | ND | Yes | 0 | 0 | 0.42 ± 0.00 | Reticular | >1,000 |
| E120D/R121E | + | - | ND | ND | 0 | 0 | 0.55 ± 0.06 | L reticular | >1,000 |
| R121S | + | - | 49 ± 8 | Yes | ND | ND | 0.48 ± 0.13 | Reticular | 340 |
| R121Q | + | - | 55 ± 5 | Yes | 0 | ND | 0.54 ± 0.07 | Reticular | 300 |
| L118E | + | - | 100 | Yes | 0 | 10 | 0.4 ± 0.03 | Reticular | 240 |
| W117E | + | - | 60 | Yes | 6 | 0* | 0.43 ± 0.08 | Reticular | 300 |
| R121E | - | - | 100 | Yes | 0 | 0* | 0.43 ± 0.05 | Reticular | >1,000 |
| R121D | - | - | 100 | Yes | 0 | 0* | 0.55 ± 0.06 | Reticular | 173 |
Restriction was measured by infecting cells expressing the indicated TRIM5αrh variants with HIV-1 and N-MLV expressing the GFP protein. After 48 h, the percentage of GFP-positive cells (infected cells) was determined by flow cytometry. For HIV-1: ++++, 100% restriction; +++, ∼75% restriction; ++, ∼50% restriction; +, ∼25% restriction; and −, <25% or no restriction. For N-MLV: ++, ∼50% restriction; +, ∼25% restriction; and −, <25% restriction. Experiments were performed at least three times, and typical results are shown.
HIV-1 reverse transcription was measured by real-time PCR 6 h after infection as described in Materials and Methods. The number represents the percentage of late reverse transcripts observed in cells expressing the indicated TRIM5αrh variant relative to the level of late reverse transcripts in control HIV-1-infected cells transduced with the empty LPCX vector. Experiments were performed at least two times.
Dimerization of TRIM5αrh variants was measured by the use of the cross-linker EGS as described in Materials and Methods.
Each TRIM5αrh variant was assayed for higher-order association with wild-type TRIM5αrh (association with wild type) or with itself (self-association), as described in Materials and Methods. In the case of association with wild-type TRIM5αrh, the percentage represents the fraction of the TRIM5αrh variant coprecipitated with wild-type TRIM5αrh relative to the amount of wild-type TRIM5αrh coprecipitated with itself. For self-association, the percentage represents the fraction of the TRIM5αrh variant coprecipitated with itself relative to the coprecipitation of wild-type TRIM5αrh with itself. *, A high background was observed for the TRIM5αrh variant in the control sample without a TRIM5α target protein. The reported values may be less reliable as a result of this background.
Binding to the HIV-1 capsid complexes was determined for each TRIM5αrh variant as described in Materials and Methods. Binding is expressed as the amount of the TRIM5αrh variant bound to HIV-1 capsid complexes divided by the amount of bound wild-type TRIM5αrh at a similar input level. Experiments were repeated at least three times. Note that, because the binding ratios are calculated at input levels at which some binding of the mutant TRIM5αrh protein to the HIV-1 capsid complexes can be detected, these ratios overestimate the relative capsid-binding affinities of the mutant proteins.
The localization of each TRIM5αrh variant was determined as described in Materials and Methods. For each TRIM5αrh variant, cytoplasmic bodies were counted for five independent cells and compared to the number in cells expressing wild-type TRIM5αrh. “L” means fewer cytoplasmic bodies than wild-type TRIM5αrh; “M” means more cytoplasmic bodies than the wild type, and “reticular” indicates a diffuse, reticular pattern of staining throughout the cytoplasm.
The half-life of each TRIM5αrh variant was estimated by pulse-chase experiments. Every experiment was repeated at least two times.
ND, not determined.
Retroviral restriction by TRIM5αrh B-box 2 mutants.
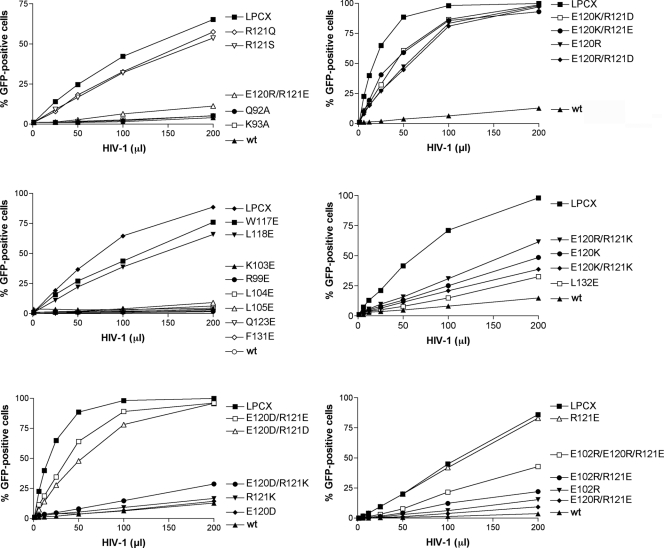
To examine the ability of the TRIM5αrh B-box 2 mutants to inhibit retroviral infection, Cf2Th cells expressing these mutants were challenged with recombinant HIV-1 expressing GFP. The TRIM5αrh mutants exhibited a range of HIV-1-restricting abilities (Fig. 4), and are rank-ordered in Table 2 according to restriction potency. Substitution of an acidic residue for arginine 121 (R121E and R121D) completely eliminated the ability of TRIM5αrh to inhibit HIV-1 infection, as has been previously seen (10, 32). Of note, some changes in the glutamic acid residues at 102 and 120, which flank the hydrophobic patch in the cluster 1 region, restored various degrees of HIV-1-restricting activity to the R121D or R121E mutants (Table 2). For TRIM5αrh mutants with an acidic residue at position 121, an arginine substitution for glutamic acid 120 resulted in the best improvement in HIV-1 restriction; lysine and aspartic acid substitutions at residue 120 were less effective. An arginine substitution for E102 was as effective as an arginine substitution for E120 in restoring HIV-1-restricting activity to TRIM5αrh variants with the R121E change. Thus, while the presence of three negatively charged residues at positions 102, 120, and 121 was associated with only a low level of TRIM5αrh function, arginine substitutions at position 102 or 120 could partially compensate for the presence of an acidic residue at position 121.
FIG. 4.
Restriction of HIV-1 infection by TRIM5αrh mutants. Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. Control cells were transduced with the empty LPCX vector. Stably expressing cell lines were selected with 5 μg of puromycin/ml. The cells were incubated with HIV-1-GFP, and the percentage of GFP-positive cells was measured 48 h later by FACS. The results of two independent experiments were similar; the results of a single experiment are shown.
The TRIM5αrh mutants that retained the wild-type arginine or a lysine residue at position 121 exhibited HIV-1-restricting ability, regardless of the particular amino acid at residue 120. However, the wild-type glutamic acid and aspartic acid residues at position 120 generally allowed the most potent HIV-1 restriction in this case. The alteration of glutamic acid 102 to arginine resulted in only a slight decrease in the potency of HIV-1 restriction when the wild-type arginine 121 was retained on TRIM5αrh. These results indicate that the nature of the charged residues that rim the hydrophobic center of cluster 1 influence the efficiency of restriction by TRIM5αrh.
Of the hydrophobic B-box 2 residues that were altered, changes in W117 and L118, in the cluster 1 hydrophobic patch, exerted the greatest impact on HIV-1-restricting activity. Thus, the TRIM5αrh B-box 2 residues important for HIV-1 restriction (arginine 121, leucine 118, and tryptophan 117) occupy a localized area on the domain surface.
The TRIM5αrh B-box 2 mutants exhibited a range of HIV-1-restricting activities. The ability of the mutants to restrict N-MLV infection was examined. The wild-type TRIM5αrh protein exhibited a modest level of restriction against N-MLV (Table 2). Whereas a few of the B-box 2 mutants that exhibited wild-type anti-HIV-1 activity retained the ability to inhibit N-MLV weakly, most of the mutants were devoid of detectable N-MLV-restricting ability. Thus, several of the mutants, including R121K, exhibited a conditional phenotype, restricting HIV-1 potently but exhibiting no inhibitory activity against the more weakly restricted N-MLV.
Inhibition of HIV-1 reverse transcription by TRIM5αrh mutants.
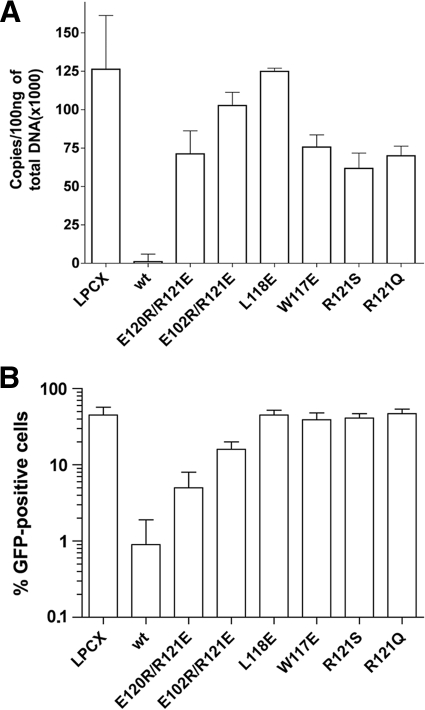
Potent HIV-1 restriction by TRIM5αrh occurs prior to the initiation of reverse transcription (22, 50). To examine the ability of the TRIM5αrh B-box 2 mutants to block HIV-1 reverse transcription, we assayed the level of late reverse transcripts in mutant-expressing cells challenged with HIV-1-GFP. The percentage of GFP-positive cells was assessed in parallel to provide an indicator of successful infection (Fig. 5). HIV-1 reverse transcripts were not detected in any of the cells expressing potently restricting TRIM5αrh B-box 2 mutants (Table 2). In contrast, in cells expressing B-box 2 mutants that did not restrict HIV-1 infection, the levels of HIV-1 reverse transcripts were similar to those in control cells not expressing TRIM5αrh (Table 2). Intermediate levels of reverse transcripts were found in most cells expressing TRIM5αrh mutants with modest HIV-1-restricting ability (Table 2). Thus, for this panel of TRIM5αrh B-box 2 mutants, the levels of HIV-1 late reverse transcripts were lowest when the degree of restriction was most potent.
FIG. 5.
Blockade of HIV-1 reverse transcription by the TRIM5αrh B-box 2 mutants. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αrh proteins or containing the empty LPCX vector were challenged at a multiplicity of infection of 0.4 with DNase-pretreated HIV-1-GFP. After 6 h, the cells were lysed, and the total DNA was extracted. (A) The levels of viral DNA were measured by quantitative real-time PCR, using a probe against GFP, as described in Materials and Methods. The means and standard deviations of the results derived from three independent experiments performed in triplicate are shown. (B) In some cases, infections were left to proceed for 48 h, and infectivity was measured as the percentage of GFP-positive cells by FACS. The values shown represent the means and standard deviations of the results obtained in three independent experiments.
Effect of TRIM5α B-box 2 changes on HIV-1 capsid binding.
Based on early, qualitative assays measuring the binding of TRIM5α mutants to HIV-1 capsid complexes, an effector function was proposed for the TRIM5α B-box 2 domain (10). Subsequently, when more quantitative capsid-binding assays were utilized, the effects of B-box 2 changes on capsid binding were recognized (32, 33). Because the expression of some TRIM5α B-box 2 mutants is elevated compared to that of the wild-type protein (9), the input levels of TRIM5α variants must be adjusted to compare capsid-binding abilities accurately. We tested the hypothesis that the major function of the TRIM5α B-box 2 domain in retrovirus restriction is to increase the avidity of capsid binding. To this end, the ability of the panel of TRIM5αrh B-box 2 mutants to bind HIV-1 CA-NC complexes assembled in vitro was assessed. The level of each TRIM5αrh variant in the cell lysate was first measured, and each lysate was diluted with control cell lysates so that comparable levels of TRIM5α proteins were present in each input sample. The adjusted cell lysates containing the TRIM5αrh variants were then serially diluted and incubated with assembled HIV-1 CA-NC complexes. The mixtures were layered on a 70% sucrose cushion and centrifuged. The amount of TRIM5αrh protein that cosedimented with the HIV-1 CA-NC complexes was measured. Comparison of the input and bound TRIM5αrh levels for each mutant with those of the wild-type TRIM5αrh protein allowed an estimation of the relative capsid-binding affinity of the mutants (Fig. 6). The relative capsid-binding ability of each mutant was calculated at the lowest input concentration of the mutant protein that yielded a detectable amount of capsid binding, thus allowing a comparison among the mutants (Fig. 7A and Table 2). A strong correlation (rs = 0.8421, P < 0.001) between binding to HIV-1 capsid complexes and HIV-1 restriction was observed for the panel of TRIM5αrh variants (Fig. 7B). These results support the hypothesis that the enhancement of capsid-binding avidity represents a major contribution of the TRIM5α B-box 2 domain to retroviral restriction.
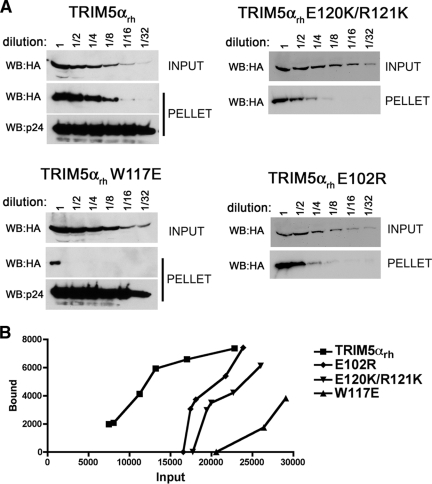
FIG. 6.
Binding of TRIM5αrh B-box 2 mutants to assembled HIV-1 capsids. (A) 293T cells were transfected with plasmids expressing the indicated wild-type and mutant TRIM5αrh proteins tagged with HA epitopes. At 36 h after transfection, the cells were lysed. Serial dilutions of the cleared lysates were made by mixing with cleared lysates from untransfected 293T cells. The lysates were incubated at room temperature for 1 h with HIV-1 CA-NC complexes that had been assembled in vitro. The mixtures were applied to a 70% sucrose cushion and centrifuged. Input represents the mixtures analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted for the HA tag. The pellet from the 70% cushion (pellet) was analyzed by Western blotting with antibodies against the HA tag and HIV-1 CA-NC protein. (B) The Western blots shown in panel A were quantitated as described in Materials and Methods. The amounts of TRIM5αrh protein in the input and pellet (bound) fractions are shown in arbitrary units.
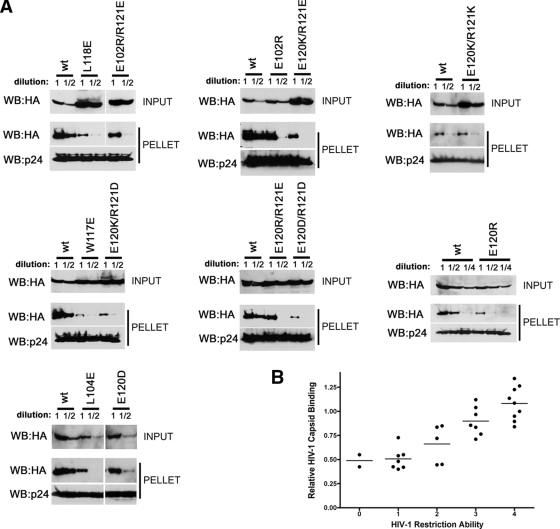
FIG. 7.
Binding of additional TRIM5αrh mutants to assembled HIV-1 capsid complexes. (A) The binding of the indicated TRIM5αrh variants to HIV-1 CA-NC complexes assembled in vitro were assessed as described in the Fig. 6 legend and Materials and Methods. (B) The relative capsid-binding ability and HIV-1-restricting activity of each TRIM5αrh B-box 2 variant were assessed as described in the Table 2 footnotes and in Materials and Methods. The means for the relative capsid-binding abilities associated with each HIV-1 restriction group are indicated by horizontal bars. The Spearman rank correlation coefficient, rs, is 0.8421, with a 95% confidence interval of 0.779 to 0.905 (two-sided P value of <0.001).
Some of the B-box 2 mutants were expressed at higher steady-state levels than the wild-type TRIM5αrh protein (see Fig. S1 in the supplemental material). Although this variable is compensated for by adjusting the input levels of TRIM5αrh in the capsid-binding assay, varying expression levels could influence the assessment of HIV-1 restriction potency. Thus, we reanalyzed the data in Fig. 7B, separating the TRIM5αrh mutants into those that were highly expressed and those expressed at wild-type TRIM5αrh levels. The strength of the correlation was not improved by taking expression level into consideration (data not shown). Thus, TRIM5αrh overexpression only minimally affects HIV-1 restriction potency in this context.
Higher-order association of TRIM5αrh B-box 2 mutants.
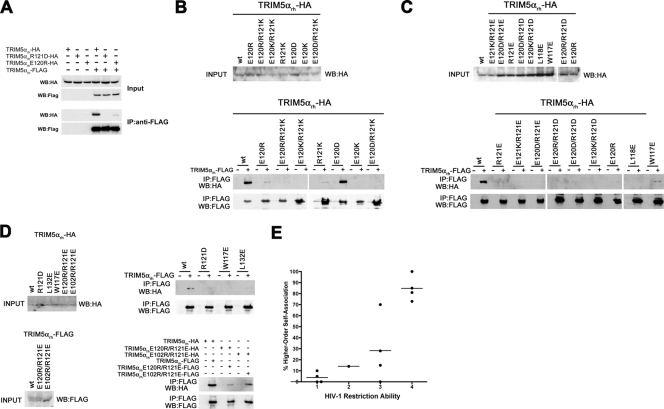
The B-box 2 domain has been shown to be important for the ability of TRIM5α dimers to associate in higher-order complexes (32). To test the ability of the B-box 2 mutants to form higher-order complexes with the wild-type TRIM5αrh proteins, cell lysates containing FLAG-tagged wild-type TRIM5αrh and HA-tagged mutant TRIM5αrh proteins were mixed. After precipitation with an anti-FLAG antibody, the precipitates were Western blotted with antibodies directed against the FLAG and HA epitope tags. The wild-type TRIM5αrh-HA protein was efficiently coprecipitated by the anti-FLAG antibody (Fig. 8), which is consistent with the ability of wild-type TRIM5αrh to self-associate in higher-order complexes. Of the TRIM5αrh mutants tested, only E120D was able to associate efficiently with wild-type TRIM5αrh (Fig. 8 and Table 2). Apparently, the ability of TRIM5αrh to form higher-order hetero-oligomers with the wild-type TRIM5αrh protein is very sensitive to changes in different surface residues of the B-box 2 domain.
FIG. 8.
Higher-order self-association of TRIM5αrh B-box 2 mutants. 293T cells were transfected with plasmids expressing the indicated wild-type or mutant TRIM5αrh proteins with a FLAG or an HA epitope tag. Cells expressing wild-type and mutant TRIM5αrh proteins were lysed 48 h after transfection. (A to D) The cell lysates were mixed, and the indicated mixtures were used for immunoprecipitation by an antibody directed against the FLAG epitope, as described in Materials and Methods. Elution of the immunocomplexes was performed with a FLAG tripeptide and analyzed by Western blotting with anti-HA and anti-FLAG antibodies. WB, Western blot; IP, immunoprecipitation. (E) The higher-order self-association and HIV-1-restricting ability, determined as described in the Table 2 footnotes and in Materials and Methods, of each TRIM5αrh variant were compared. The means of the values for higher-order self-association within each restriction group are indicated by horizontal bars. The Spearman rank correlation coefficient, rs, is 0.862, with a 95% confidence interval of 0.665 to 1.0 (two-sided P value of <0.001).
We examined the ability of a subset of TRIM5αrh mutants to self-associate into higher-order homo- oligomers (Fig. 8D and see Fig. S4 in the supplemental material). TRIM5αrh mutants that exhibited efficient ability to form higher-order homo- oligomers restricted retrovirus infection with modest or high efficiency (Table 2). Conversely, most of the weakly restricting TRIM5α mutants either did not self-associate into higher-order homo-oligomers or did so weakly. Thus, there is a significant correlation between homologous higher-order self-association of the TRIM5αrh variants and HIV-1-restricting ability (rs = 0.862, P < 0.001) (Fig. 8E). This observation is consistent with a model in which homologous higher-order self-association contributes to the HIV-1-restricting ability of TRIM5α.
Cellular localization of TRIM5αrh B-box 2 mutants.
Changes in the B-box 2 domain can change the cytoplasmic staining of the overexpressed TRIM5αrh protein from cytoplasmic bodies to a diffuse, reticular pattern (10). Although a few of the B-box 2 mutants localized in well-defined cytoplasmic bodies, most of the mutants exhibited reticular staining patterns (see Fig. S5 in the supplemental material and Table 2). These results support previous studies that suggested that cytoplasmic body formation is not necessary for HIV-1 restriction by TRIM5αrh (10, 40, 49).
DISCUSSION
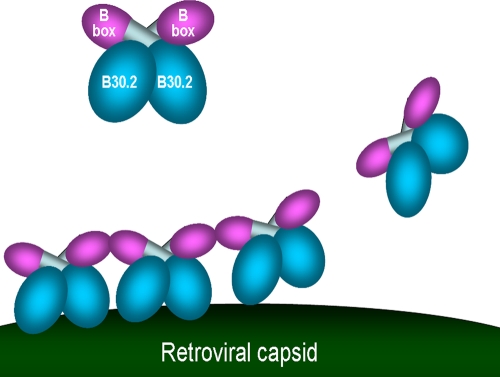
We report here the structure of the TRIM5α B-box 2 domain, which reveals its unusual surface properties. We utilize this information to investigate which of the many reported functions of this domain relate to retroviral restriction. The TRIM5α B-box 2 ββα fold is similar to that of other B-box 2 and RING-like domains. One notable feature is the surface exposure of hydrophobic residues, which are organized into two clusters, cluster 1 and cluster 2. We provide evidence that the B-box 2 domain contributes to the self-association of TRIM5α proteins. A stronger, zinc-mediated interaction contributed to the formation of dimers between isolated TRIM5α B-box 2 domains. Since the TRIM5α B-box 2 domain is not required for dimerization (20) and free zinc ion concentrations in the cytoplasm are low, this zinc-mediated interaction is unlikely to contribute to the association of the subunits in the TRIM5α dimer. All of the TRIM5αrh B-box 2 mutants studied herein efficiently dimerized. We identified a second, weaker interaction between the B-box 2 domains involving residues near or within cluster 1. Cluster 1 is likely exposed on the surface of the TRIM5α dimer, since it apparently contributes to the higher-order self-association of TRIM5α proteins. Alteration of W117, L118 and R121 within or near Cluster 1 resulted in TRIM5αrh proteins that dimerized, but could not further associate to form higher-order oligomers. Higher-order self-association of the panel of B-box 2 mutants correlated with the ability of TRIM5αrh to restrict HIV-1 infection. The formation of short-range, higher-order TRIM5α arrays might contribute to capsid-binding avidity (Fig. 9). If the long-range geometry of these TRIM5α arrays is incompatible with that of the retroviral capsid, higher-order self-association could promote the dissolution of the viral capsid during the restriction process (9, 41, 51).
FIG. 9.
Model for the role of the TRIM5α B-box 2 domain in HIV-1 restriction. The TRIM5αrh dimer is depicted, with the B-box 2 domains (magenta) and B30.2(SPRY) domains (blue) highlighted. Interactions involving the cluster 1 region of the B-box 2 domain increase the avidity of TRIM5αrh for the retroviral capsid, promoting the aggregation of the TRIM5αrh protein on the capsid surface.
Specific changes in or near cluster 1 were shown to decrease the ability of TRIM5αrh to bind HIV-1 capsid-like complexes. The TRIM5α B-box 2 mutants exhibited a wide range of HIV-1-restricting abilities, affording us the opportunity to test the hypothesis that the major contribution of the B-box 2 domain to TRIM5α restriction function is mediated through capsid binding. The strong correlation that we observed between capsid-binding ability and HIV-1 restriction potency supports this hypothesis. Notably, we did not identify any B-box 2 mutants that bound the HIV-1 capsid complexes efficiently but failed to restrict HIV-1 infection; in other words, no candidate for a B-box 2 mutant defective in an “effector” function was apparent in this sizable mutant panel. Collectively, more than 80% of the TRIM5α B-box 2 domain surface has been altered (10, 33), increasing the likelihood that the major functions of this domain have already been uncovered.
Coprecipitation assays were used to assess the ability of TRIM5αrh mutant dimers to associate with wild-type TRIM5αrh dimers (higher-order heterologous association) or with themselves (higher-order homologous association). The higher-order homologous association of TRIM5αrh mutants correlated with HIV-1-restricting activity better than heterologous association. This is expected, because in the cells tested for restriction activity, only mutant TRIM5αrh proteins were present. Some of the cluster 1 mutants formed higher-order homologous associations better than heterologous associations, indicating that the introduced changes were self-compatible but disruptive of interactions with the wild-type TRIM5αrh protein. This observation supports the importance of the B-box 2 surface proximal to cluster 1 in mediating the intermolecular contacts that stabilize the higher-order self-association of TRIM5α dimers.
The hydrophobic cluster 1 surface of the TRIM5αrh B-box 2 domain is dominated by the indole ring of W117 (W115 in human TRIM5). W117, along with L118 (L116 in human TRIM5), appears to contribute significantly to the higher-order self-association, capsid-binding avidity and HIV-1-restricting ability of TRIM5αrh. In addition, a basic residue, preferably an arginine residue, flanking this hydrophobic patch contributes to these functions; an arginine at the equivalent position of human TRIM5α has previously been shown to contribute to N-MLV restriction (10). Some flexibility with respect to the position of this arginine residue appears to be tolerated in TRIM5αrh, with arginine at positions 102 or 120 able to compensate for the alteration of R121. The W117, L118, and R121 residues are all conserved in TRIM6 and TRIM34, whose B-box 2 domains can functionally substitute for those of TRIM5 (31). These residues diverge in more distant TRIM proteins such as TRIM21, for example; the RING domain, but not the RING and B-box 2 domains, of TRIM5αrh can be functionally replaced by those of TRIM21 (31). Although TRIM6 and TRIM34 exhibit at best marginal ability to restrict retrovirus infection (30, 60), higher-order self-association might contribute to other functions of these proteins. In this respect, it is notable that the dimeric contacts in the crystallized MuRF1 (TRIM63) B-box domain correspond to the cluster 1 surface of TRIM5 (37). Thus, many TRIM proteins may utilize the corresponding surface of the B-box domain to modulate protein-protein interactions, including self-association.
The levels of HIV-1 reverse transcripts detected in the cells expressing the TRIM5αrh mutants after HIV-1 challenge were lowest when infection was potently restricted. Conversely, in cells expressing the most restriction-defective TRIM5αrh proteins, the levels of reverse transcripts were comparable to those in control cells lacking TRIM5αrh. In cells expressing TRIM5αrh mutants with intermediate levels of restriction potency, a wide range of reverse transcript levels were observed following HIV-1 challenge. These results suggest that TRIM5αrh restriction of HIV-1 occurs early in the infection process, precluding reverse transcription when potent and allowing various degrees of reverse transcription when less potent.
Several TRIM5αrh B-box 2 did not form cytoplasmic bodies but still exhibited fairly potent restriction of HIV-1 infection. This observation is consistent with previous studies that suggested that cytoplasmic body formation by TRIM5α is not required for retroviral restriction (40, 49). Our data do not rule out the possibility that the B-box 2-mediated higher-order self-association of TRIM5α is required for cytoplasmic body formation; however, higher-order self-association appears not to be sufficient for cytoplasmic body formation.
As previously observed (10), some changes in the B-box 2 domain can increase the longevity of the relatively short-lived TRIM5α protein. Our data confirm that rapid turnover is not required for the HIV-1-restricting activity of TRIM5α; no correlation between the half-lives and retrovirus-inhibiting abilities of the TRIM5αrh B-box 2 mutants was apparent. Our data also indicate that cytoplasmic body formation is not necessary for the rapid turnover of TRIM5α.
The importance of B-box 2-mediated higher-order self-association and capsid-binding avidity to retroviral restriction should motivate efforts to understand the TRIM5α and capsid structures that contribute to these processes. Such understanding may suggest new interventional approaches to HIV-1 infection.
Supplementary Material
Acknowledgments
We thank Takashi Umehara for analytical ultracentrifugation and dynamic light scattering measurements and useful discussions and Toshio Nagashima for molecular dynamics calculation of the association model and useful discussions. We also thank Satoru Watanabe, Takushi Harada, Takeshi Nagir, Yasuko Tomo, Masaomi Ikari, Kazuharu Hanada, Yukiko Fujikura, and Akiko Tanaka for sample preparation and showing the screening data of B-box 2 domains. We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
We acknowledge the National Institutes of Health (grants AI063987 and AI076094 and a Center for AIDS Research Award AI60354), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper for research funding. F.D.-G. is a recipient of a K99/R00 Pathway to Independence Award from the National Institutes of Health (1K99MH086162-01), an American Foundation for AIDS Research Mathilde Krim fellowship in basic biomedical research (106987-43-RFHF), and a Claudia Adams Barr award from the Dana-Farber Cancer Institute. The B-box 2 structure determination was supported by the Riken Structural Genomics/Proteomics Initiative of the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 5 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ayed, A., F. A. Mulder, G. S. Yi, Y. Lu, L. E. Kay, and C. H. Arrowsmith. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8:756-760. [DOI] [PubMed] [Google Scholar]
- 1a.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 2.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, G., Y. Kozyrev, and S. L. Hu. 2008. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. USA 105:3569-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzovic, P. S., P. Rajagopal, D. W. Hoyt, M. C. King, and R. E. Klevit. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8:833-837. [DOI] [PubMed] [Google Scholar]
- 6.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 7.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2002. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J. Virol. 76:12866-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz-Griffero, F., A. Kar, M. Lee, M. Stremlau, E. Poeschla, and J. Sodroski. 2007. Comparative requirements for the restriction of retrovirus infection by TRIM5α and TRIMCyp. Virology 369:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Griffero, F., A. Kar, M. Perron, S. H. Xiang, H. Javanbakht, X. Li, and J. Sodroski. 2007. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5α B-box 2 domain. J. Virol. 81:10362-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349:300-315. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Griffero, F., N. Vandegraaff, Y. Li, K. McGee-Estrada, M. Stremlau, S. Welikala, Z. Si, A. Engelman, and J. Sodroski. 2006. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 351:404-419. [DOI] [PubMed] [Google Scholar]
- 13.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 14.Ganser-Pornillos, B. K., U. K. von Schwedler, K. M. Stray, C. Aiken, and W. I. Sundquist. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guntert, P. 2009. Automated structure determination from NMR spectra. Eur. Biophys. J. 38:129-143. [DOI] [PubMed] [Google Scholar]
- 16.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennig, J., A. Bresell, M. Sandberg, K. D. Hennig, M. Wahren-Herlenius, B. Persson, and M. Sunnerhagen. 2008. The fellowship of the RING: the RING-B-box linker region interacts with the RING in TRIM21/Ro52, contains a native autoantigenic epitope in Sjogren syndrome, and is an integral and conserved region in TRIM proteins. J. Mol. Biol. 377:431-449. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, T., P. Guntert, and K. Wuthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209-227. [DOI] [PubMed] [Google Scholar]
- 19.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 20.Javanbakht, H., W. Yuan, D. F. Yeung, B. Song, F. Diaz-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234-246. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, B. A. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313-352. [DOI] [PubMed] [Google Scholar]
- 22.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kentsis, A., and K. L. Borden. 2004. Physical mechanisms and biological significance of supramolecular protein self-assembly. Curr. Protein Pept. Sci. 5:125-134. [DOI] [PubMed] [Google Scholar]
- 24.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 99:15404-15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kigawa, T., T. Yabuki, N. Matsuda, T. Matsuda, R. Nakajima, A. Tanaka, and S. Yokoyama. 2004. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics 5:63-68. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, N., J. Iwahara, S. Koshiba, T. Tomizawa, N. Tochio, P. Guntert, T. Kigawa, and S. Yokoyama. 2007. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 39:31-52. [DOI] [PubMed] [Google Scholar]
- 27.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph 14:29-32; 51-55. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., S. Koshiba, F. Hayashi, N. Tochio, T. Tomizawa, T. Kasai, T. Yabuki, Y. Motoda, T. Harada, S. Watanabe, M. Inoue, Y. Hayashizaki, A. Tanaka, T. Kigawa, and S. Yokoyama. 2008. Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode. J. Biol. Chem. 283:27165-27178. [DOI] [PubMed] [Google Scholar]
- 30.Li, X., B. Gold, C. O'Huigin, F. Diaz-Griffero, B. Song, Z. Si, Y. Li, W. Yuan, M. Stremlau, C. Mische, H. Javanbakht, M. Scally, C. Winkler, M. Dean, and J. Sodroski. 2007. Unique features of TRIM5α among closely related human TRIM family members. Virology 360:419-433. [DOI] [PubMed] [Google Scholar]
- 31.Li, X., Y. Li, M. Stremlau, W. Yuan, B. Song, M. Perron, and J. Sodroski. 2006. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5α (TRIM5α) by heterologous TRIM domains. J. Virol. 80:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X., and J. Sodroski. 2008. The TRIM5α B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82:11495-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, X., B. Song, S. H. Xiang, and J. Sodroski. 2007. Functional interplay between the B-box 2 and the B30.2(SPRY) domains of TRIM5α. Virology 366:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massiah, M. A., J. A. Matts, K. M. Short, B. N. Simmons, S. Singireddy, Z. Yi, and T. C. Cox. 2007. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J. Mol. Biol. 369:1-10. [DOI] [PubMed] [Google Scholar]
- 35.Massiah, M. A., B. N. Simmons, K. M. Short, and T. C. Cox. 2006. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol. 358:532-545. [DOI] [PubMed] [Google Scholar]
- 36.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mrosek, M., S. Meier, Z. Ucurum-Fotiadis, E. von Castelmur, E. Hedbom, A. Lustig, S. Grzesiek, D. Labeit, S. Labeit, and O. Mayans. 2008. Structural analysis of B-Box 2 from MuRF1: identification of a novel self-association pattern in a RING-like fold. Biochemistry 47:10722-10730. [DOI] [PubMed] [Google Scholar]
- 38.Newman, R. M., L. Hall, A. Kirmaier, L. A. Pozzi, E. Pery, M. Farzan, S. P. O'Neil, and W. Johnson. 2008. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 4:e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 81:2138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poyurovsky, M. V., C. Priest, A. Kentsis, K. L. Borden, Z. Q. Pan, N. Pavletich, and C. Prives. 2007. The Mdm2 RING domain C terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 26:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawyer, S. L., M. Emerman, and H. S. Malik. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 48.Schweiger, S., and R. Schneider. 2003. The MID1/PP2A complex: a key to the pathogenesis of Opitz BBB/G syndrome. Bioessays 25:356-366. [DOI] [PubMed] [Google Scholar]
- 49.Song, B., F. Diaz-Griffero, D. H. Park, T. Rogers, M. Stremlau, and J. Sodroski. 2005. TRIM5α association with cytoplasmic bodies is not required for antiretroviral activity. Virology 343:201-211. [DOI] [PubMed] [Google Scholar]
- 50.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 51.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao, H., B. N. Simmons, S. Singireddy, M. Jakkidi, K. M. Short, T. C. Cox, and M. A. Massiah. 2008. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 47:2450-2457. [DOI] [PubMed] [Google Scholar]
- 54.Torok, M., and L. D. Etkin. 2001. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 67:63-71. [DOI] [PubMed] [Google Scholar]
- 55.Virgen, C. A., Z. Kratovac, P. D. Bieniasz, and T. Hatziioannou. 2008. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. USA 105:3563-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson, S. J., B. L. Webb, L. M. Ylinen, E. Verschoor, J. L. Heeney, and G. J. Towers. 2008. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. USA 105:3557-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human TRIM5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 59.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Pt. A):99-112. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, F., T. Hatziioannou, D. Perez-Caballero, D. Derse, and P. D. Bieniasz. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396-409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.