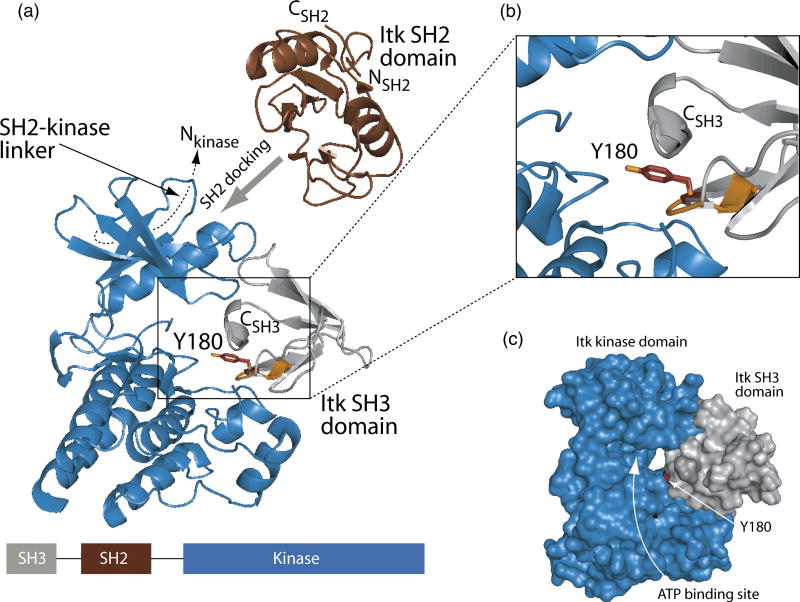
Figure 6. Model of in cis Itk autophosphorylation.
(A) The Itk kinase domain (shown in blue, PDB ID 1SNX) was aligned with the structure of the substrate bound insulin receptor kinase domain (PDB ID 1IR3). The insulin receptor kinase domain itself is not shown for clarity while the tyrosine containing peptide substrate in that structure is indicated in orange. Itk Y180 (represented by sticks in red) within the Itk SH3 domain (grey ribbons) was placed into the active site of the kinase domain by superimposing Y180 (red) with the tyrosine residue of the insulin receptor kinase substrate (orange). To achieve efficient phosphorylation of Y180 in the SH3 domain, the SH2 domain must dock onto the Itk kinase domain outside of the active site19. Additionally, the 17-amino acid SH2-kinase linker contacts the ‘backside’ of the small lobe of the kinase domain (indicated with dashed line)18. Thus, restraints imposed by the amino and carboxy termini of the SH2 domain and its neighboring domains (SH3 and kinase) suggest that SH2 docking likely occurs on the small lobe of the kinase domain (indicated with wide grey arrow). The linear domain structure of the SH3-SH2-kinase portion of Itk is shown at the bottom of the figure. The enlarged view shown in (B) more clearly shows the superposition of Y180 with the peptide substrate tyrosine of the insulin receptor kinase structure. (C) Surface rendering of the Itk kinase domain (blue) and the Itk SH3 domain (grey). Y180 is red and the ATP binding cleft of the Itk kinase domain is labeled. Figures 5 and 6 were generated using PyMol40.