Abstract
Retrogenes are processed copies of genes that are inserted into new genomic regions and that acquire new regulatory elements from the sequences in their surroundings. Here we use a comparative approach of phylogenetic footprinting and a non-comparative approach of measuring motif over-representation in retrogenes in order to describe putative elements present in cis-regulatory regions of 94 retrogenes recently described in Drosophila. The detailed examination of the motifs found in the core promoter regions of retrogenes reveals an abundance of the DNA replication-related element (DRE), the Initiator (Inr), and a new over-represented motif that we call the GCT motif. Parental genes also show an abundance of DRE and Inr motifs, but these do not seem to have been carried over with retrogenes. In particular, we also examined motifs upstream of retrogenes expressed in adult testis and were able to identify 6 additional over-represented motifs. Comparative analyses provide data on the conservation and origin of some of these motifs and reveal 15 additional conserved motifs in these retrogenes. Some of those conserved motifs are sequences bound by known transcription factors, while others are novel motifs. In this report we provide the first genome-wide data on which specific cis-regulatory regions can be recruited by retrogenes after they are inserted into new coding regions in the genome. Future experiments are needed to determine the function and role of the new elements presented here.
Keywords: retrogene, Drosophila, testis expression, regulatory motifs
INTRODUCTION
When an organism’s mRNA is reverse-transcribed and inserted into the genome, the result is a processed copy of a gene referred to as a retrogene [1]. The retrogene is therefore an example of gene duplication that does not contain introns or cis-regulatory regions. Since these processed copies lack regulatory regions, they will often degenerate becoming pseudogenes [2]. Yet many processed gene copies are known to produce functional proteins, often in the male germline of flies and mammals [3; 4; 5; 6; 7]. How a retrogene acquires its regulatory elements from the target site of insertion is a major question in efforts to understand the origin of new genes.
We have recently examined different mechanisms to explain how retrogenes acquire male germline expression [6]. We showed that retrogenes do not generally carry regulatory regions from aberrant or normal transcripts of their parental genes, and that expression patterns of the closely neighboring genes are not consistently shared with retrogenes. We also reported that transposable elements do not appear to frequently contribute regulatory regions to retrogenes. Interestingly, we found that there is an excess of retrogenes in male germline neighborhoods, and this cannot be explained simply by the reported insertional biases of the retroelement machinery used for retroposition [8]. Rather, we believe that these results reflect selection in favor of retrogenes inserted in germline regions and selection at the sequence level, which results in testis expression.
Transcription requires the presence of cis-regulatory motifs, including promoter motifs and other cis-regulatory regions (enhancers and silencers) [9]. Here we study the cis-regulatory regions of retrogenes; in particular, we use computational methods to analyze 94 retroposed genes recently described in Drosophila [3] for the presence or absence of known promoter motifs and new over-represented motifs. Given our previous analysis of the presence/absence of these retrogene set in 11 other Drosophila species [3], we used comparative and non-comparative approaches here to describe the elements present in cis-regulatory regions of these Drosophila retrogenes. Little information exists about how new genes recruit new cis-regulatory regions. We provide the first genome-wide data on the putative quality of cis-regulatory regions that are recruited after the insertion of a new coding region in the genome.
This detailed examination of the motifs found in the core promoter regions of retrogenes (i.e. from −100 to +40 in the nucleotide sequence) revealed an abundance of the DNA replication-related element (DRE), the Initiator (Inr) motif, and a new over-represented motif that we call the GCT motif. We also examined the existence of over-represented motifs in retrogenes expressed in adult testis. To include testis enhancers, we investigated a larger region (from −1000 to +40 in the sequence) and identified six additional over-represented sequences. We use comparative analyses to shed light on the conservation and origin of some of these sequences. In addition, we identified 15 putative conserved motifs in retrogenes. Some of those conserved motifs are similar to sequences that are known to be bound by transcription factors, while others are novel sequences.
RESULTS
TSS identification and quality check
Mononucleotide and CA dinucleotide distribution surrounding transcriptional start sites
Correct positioning of a gene’s transcriptional start site (TSS) is essential for analyzing the structure of promoters and motifs [10]. The quality of the TSS determinations for the genes under study is assessed by the mononucleotide and CA dinucleotide distribution surrounding the TSS [11]. In Drosophila, T and A have been shown to decrease in frequency upstream of the TSS, while G and C increase in frequency. This trend changes at the TSS, such that T and A increase in frequency while G and C decrease [11]. On the other hand, CA dinucleotides are expected to peak at the TSS due to the presence of the initiator motif (TCAGTY) found in ~20–29% of well characterized genes [10; 11].
Sixty-five of the 94 retrogenes and 83 of their parental genes have an annotated 5′UTR, meaning that Ensembl features a 5′UTR annotated based on EST and cDNA data [12]. We refer to these genes as annotated (see Materials and Methods for details). To explore the accuracy of this annotation, we retrieved the region between −100 and +40 nucleotides (nt) surrounding these annotated TSSs. We subsequently determined the frequency of the dinucleotide CA in this 140-nt region, and the frequency of each mononucleotide in a wider window (−2000 to +100) using a 20-nt bin. The results for the retrogenes and the parental genes are shown in Supplementary Figure 1. Based on the expected mononucleotide pattern at bin 101 (corresponding to the TSS site) and the high frequency of the CA dinucleotide close to the TSS in parental genes, the TSS is shown to be quite accurately positioned for the parental genes with a 22% frequency of CA. However, the mononucleotide pattern in the case of the retrogenes is not as expected, and the mono- and dinucleotide patterns vary significantly among the different retrogenes; seven values are between 12 and 17% in positions ranging from −51 to +20 for the CA dinucleotide distribution. We explore these deviations in more detail below.
TSS identification using the McPromoter package
A second strategy was adopted in parallel to predict the position of the TSS in all annotated genes of our data set. We retrieved a larger putative promoter region between positions −500 and +100 from “annotated” genes and predicted the TSS using the program McPromoter [13]. A value of 0.7 (on a maximum of 1.0) was chosen as the conservative sensitivity threshold to detect the putative TSSs [10]. We found significantly more hits for annotated parental genes than annotated retrogenes (Table 1; P < 10−5). McPromoter predicted the TSS for 80% of the parental genes, but only 28% of the retrogenes. The median hit scores between parental genes and retrogenes are not significantly different (Mann-Whitney U test, P = 0.2662). The predicted TSSs fall within the region between −300 and +50 at the annotated 5′ end, which is even narrower than the recommended guidelines of −1000 to +100 [10].
Table 1.
Genes with known 5′ UTRs whose TSSs were predicted by McPromoter.
| Gene type | No. predicted | No. not predicted | Ratio of predicted to total |
|---|---|---|---|
| Retrogene | 18 | 47 | 0.2769 |
| Parental gene | 66 | 17 | 0.7952 |
P < 10−5, Fisher’s exact test.
The annotated retrogene set shows deviations from the expected distributions of mononucleotides and dinucleotides in the region of the TSS, and these TSSs are predicted less often by McPromoter than for their parental genes. There are several possible explanations for these deviations: (a) retrogenes are more poorly annotated than parental genes; (b) retrogenes have fewer alternative transcripts with different TSSs, and therefore fewer chances for detection by McPromoter; or (c) retrogenes use different features in the promoter regions than parental and other genes that have been described so far.
Establishing the 5′ end of a transcript is difficult, even when working with libraries constructed from capped mRNAs [10]. It is therefore reasonable to presume that the more cDNAs or ESTs are used to define the 5′ end of a gene, the more reliable the annotation will be. Therefore, retrogenes may be more poorly annotated than parental genes if they are expressed at lower levels and therefore have fewer cDNAs or ESTs to define their transcript(s). If this is true, retrogenes should show significantly shorter 5′UTRs than parental genes and there should be a positive correlation between the number of cDNAs or ESTs and the ability of McPromoter to predict the TSS.
To test this idea, we analyzed the relationship between McPromoter predictions and the number of cDNAs or ESTs in parental genes and retrogenes. We observed no difference in the median number of cDNAs or ESTs between TSSs predicted or non-predicted in parental genes by McPromoter (medianP = 37.5000; medianNP = 38.8000; Mann-Whitney U test, P = 0.3607). However, the same analysis for retrogenes revealed a significant difference (medianP = 21.5000; medianNP = 6.0000; Mann-Whitney U Test P= 0.0109). This suggests that the number of cDNAs and ESTs (i.e., the approximate gene expression level) positively correlates with McPromoter TSS prediction in retrogenes. However, the average number of cDNAs and ESTs was very similar between non-predicted parental genes (X̄NP = 56.9686) and predicted retrogenes (X̄P = 57.1944), suggesting that this putative relationship between expression activity and the presence of TSS features in retrogenes should be examined further.
In addition, retrogenes may have fewer transcripts than parental genes and therefore fewer chances to be detected, especially if the TSSs of the different transcripts are not separated by more than 500 bp. However, we observed no difference in the median number of transcripts between predicted and non-predicted TSSs in parental genes (medianP = 1.0000; medianNP = 1.0000; Mann-Whitney U test, P = 0.1970) or retrogenes (medianP = 1.0000; medianNP = 1.0000; Mann-Whitney U test, P = 0.2500).
The median length of the 5′ UTR in the parental genes was 141 nt, compared to only 101 nt in retrogenes. This difference is significant (Mann-Whitney U test, P = 0.0012). If this difference is due to the fact that retrogene TSSs are often inappropriately annotated and lie an average of 50 bp upstream of their reported location (mean length of the 5'UTR is 198 and 147 for parental and retrogenes respectively), our promoter analysis should include a longer region upstream of the retrogene. Therefore for the McPromoter prediction, we used a longer, 500-nt region at the 5′ end that should compensate for this difference as long as there are no introns within the 5′UTR. Of the 83 annotated parental genes, 23 have an intron in the 5′UTR. The average length of the first exon in these 23 parental genes is 159 bp. Only two genes have a first exon shorter than 50 bp. This suggests that the 500-nt upstream regions that we used to run TSS predictions in McPromoter are in fact likely to contain the TSS; this region lies within ~50 nt from the region we examined in parental genes. However, the regions used for mononucleotide and CA analyses may lie an average of 50 nt from the TSS.
Poor annotation due to the number of transcripts does not seem to explain the relatively low prediction rate by McPromoter. The likely explanation is that retrogenes have different characteristics from canonical genes. One possibility is that a large number of retrogenes are young genes that have not yet picked up the canonical features. Another possibility is that retrogenes, given that they often show atypical patterns of expression [4; 6; 7], use non-canonical promoters. These could be tissue-specific promoters like the β2-tubulin gene, which can recruit RNA polymerase II and drive late spermatogenesis-specific expression in Drosophila [14]. Consistent with this view, we found several over-represented motifs in the putative promoter and cis-regulatory regions that have not previously been described (see below).
Core promoter features in retrogenes
TSS identification and quality checks were performed as described above, and they were the first step in the core promoter analysis. We subsequently examined features of the core promoter, since gene expression is ultimately regulated by the interaction between DNA motifs proximal to the TSS and the transcription factors that recognize and bind these specific sequences. Only those pairs of retrogenes and parental genes in which the parental 5′ end was annotated were used for these analyses, since these genes showed the expected distributions of mononucleotides and CA, and their TSS was predicted by McPromoter. For the data set of annotated retrogenes, genes whose TSSs were predicted by McPromoter were analyzed separately from the others. For retrogenes whose TSS was predicted, both the McPromoter TSS and the annotated TSS were analyzed separately (Table 2).
Table 2.
Motifs detected in the region between − 100 nt and + 40 nt based on both McPromoter-predicted TSS and database-annotated TSS for the retrogenes and parental genes with annotated 5′ UTRs.
| Motif Name | Motif name (Fitzgerald et al. 2006) | Motif number (Ohler et al. 2002) | Consensus sequence | Retrogenes (n = 65) | Parental Genes (n = 83) | |||
|---|---|---|---|---|---|---|---|---|
| ND (47) | D (18) |
ND (17) | D (66) | |||||
| TSS a | TSS b | |||||||
| TATA | DMp1 | 3 | TATAWAA | 8 | 2 | 2 | 2 | 8 |
| INR | DMp2 | 4 | TCAKTY | 16 | 2 | 1 | 3 | 17 |
| INR1 | DMp3 | - | TCATTCG | 1 | 1 | 1 | 1 | 0 |
| MTE | - | 10 | CSSAACGS | 2 | 0 | 0 | 0 | 3 |
| DRE | NDM4 | 2 | WATCGATW | 5 | 9 | 8 | 6 | 20 |
| - | DMv2 | 8 | TGGYAACR | 0 | 1 | 1 | 0 | 3 |
| - | DMv3 | 7 | CAYCNCTA | 2 | 2 | 2 | 1 | 13 |
| - | DMv4 | 1 | GGYCACAC | 2 | 4 | 2 | 4 | 20 |
| - | DMv5 | 6 | GTATWTTT | 3 | 0 | 0 | 0 | 8 |
| E-box | NDM5 | 5 | CAGCTSW | 1 | 5 | 5 | 1 | 7 |
| DPE | DMp4 | 9 | KCGGTTS | 0 | 0 | 0 | 0 | 2 |
| GCT | - | - | YGGCTTTK | 7 | 0 | 0 | 1 | 1 |
Total numbers of genes are given in parentheses. D refers to a TSS detected by McPromoter. ND means that the TSS was not detected by McPromoter. The high abundance of some motifs is highlighted in bold.
TSS is based on the database.
TSS is based on McPromoter prediction.
To study the occurrence of known binding sites in the regulatory regions of retrogenes and parental genes, we took advantage of recent work on genome-wide motif prediction in D. melanogaster [10; 11], which defined composition and distribution patterns of DNA motifs in gene promoters. We detected known motifs using Patser software [15] in regions from −100 to +40 of “annotated” genes (with the TSS defined as base +1). The results are reported in Table 2. Details concerning the genes with TSSs predicted by McPromoter and the position of specific motifs is given in Supplementary Table 1.
Our data for genes with a TSS predicted by McPromoter show an abundance of DRE, Inr, and motif 1 in parental genes, and an abundance of DRE and Inr in retrogenes. Motif 1 was first reported by Ohler et al. [10]. A DRE was found 14 times in retrogenes and 26 times in parental genes (Supplementary Table 1). Previous genome-wide analyses showed DRE to be an abundant motif in the promoter region of D. melanogaster genes [10; 11]. The protein that binds to the DRE has been shown to form part of a complex that has replaced the TBP in many promoters [16]. It is not surprising, therefore, that both types of genes show an abundance of the DRE motif though we do not believe that this abundance is a result of carryover from parental upstream sequences into retrogenes given our previously published results [6].
We also explored whether retrogenes carry over any downstream promoter elements from parental genes. Our results on the pairs of parental and retrogenes in which McPromoter predicted the TSS (14/18) revealed no bias for the presence of DPE or MTE; these motifs would be present in the transcript after retroposition because they are downstream promoter elements [9; 10; 17]. Out of the 14, none shows DPE or MTE in either the retrogene or parental gene (Supplementary Table 1). Consistent with previous expression and sequence analyses [6], we do not have evidence supporting the idea that the parental gene donates downstream promoter elements to the retrogene.
We explored the use of additional unknown motifs in the 47 annotated retrogenes lacking a good hit for McPromoter. As previously mentioned, the promoter regions of these genes may have unexpected features that prevent the detection of their TSSs by prediction tools that rely on relatively few motifs, such as McPromoter. Therefore the programs MEME [18] and Consensus [19] were used to detect the presence of additional, over-represented motifs in the region between −100 and +40 in the TSS. We identified a new motif, which we call GCT, that occurs in the upstream regions of seven annotated retrogenes (Table 2). The GCT motif consensus sequence, YGGCTTTK, is found at positions ranging from −65 to −11 upstream of the TSS in these seven retrogenes (see Supplementary Table 2). Further inspection revealed that this motif is likely to be directional, since it occurs only in the positive strand in these genes. We tested the over-representation of this motif by randomly sampling 500 annotated genes in the genome and comparing the ratio at which this element was found (6/500) with the ratio detected in our set of retrogenes. Fisher’s exact test clearly shows significance (P = 0.0002). Interestingly, six of the seven genes with a GCT motif in their promoter regions are expressed in the testis, suggesting that GCT motifs might help to determine tissue-specific expression in these retrogenes.
Are there any adult testis-specific motifs in retrogenes?
We therefore sought to examine in greater detail the possible role of specific motifs in regulating the testis-specific expression of several groups of genes: 48 retrogenes already shown to be expressed in adult testis [3], 14 retrogenes expressed specifically in the adult testis as revealed using EST and cDNA data (see Materials and Methods), and 34 retrogenes uniquely expressed in testis according to FlyAtlas [6; 20]. We performed a motif search using the program MEME in a region extending from 1,000 nt upstream of the TSS to 40 nt downstream of the TSS. Expression information for these genes and details of the motif analysis are given in Supplementary Tables 3 and 4.
Two over-represented motifs, with the consensus sequences TBGHYTKGGSCA and GCKCCAGYSAA, were detected, respectively, in the upstream region of 18 and 10 of the 48 retrogenes expressed in the adult testis (Table 3 and Supplementary Table 3). We refer to these motifs as testis associated 1 and 2 (TA1 and TA2). We tested their over-representation by randomly sampling 250 genes in the genome and comparing the ratio at which these motifs were found (22/250 for TA1; 1/250 for TA2). For the comparison between the observation and random samples, Fisher’s exact tests clearly indicate significance (P < 1 × 10−5 for both). Six of 18 occurrences (33%) and three out of 10 occurrences (30%) were located inside the coding region of the nearest 5′ side neighboring gene or of itself. Even excluding these cases, there is over-representation of these two motifs in the retrogenes compared to the 250 genes randomly sampled from the genome (P < 0.005 for both by Fisher’s exact tests).
Table 3.
New motifs detected upstream of retrogenes expressed in adult testis.
| # a | Motif logo b | Multilevel consensus sequence c | Occurrences | P value (Fisher’s exact test) |
|---|---|---|---|---|
| TA1 | TBGHYTKGGSCA | 18 | < 1 × 10−5 | |
| TA2 | GCKCCAGYSAA | 10 | < 1 × 10−6 | |
| TS1 | CTSWGTGCM | 7 | < 1 × 10−4 | |
| TS2 | SACMRWGSMMWG | 8 | < 1 × 10−4 | |
| TS3 | YGSMYCHTGYKGMCC | 32 | < 1 × 10−10 | |
| TS4 | CCSCTGCYSVTYCSC | 18 | < 1 × 10−10 |
Over-represented motifs detected in the data sets of 48 retrogenes with expression in adult testis (motifs TA1 and TA2),14 retrogenes expressed only in adult testis based on EST and cDNA library data (motifs TS1 and TS2), and 34 retrogenes with expressed only in the testis based on Fly Atlas data (motifs TS1 and TS2)
Degenerate bases are represented using IUPAC letters
We also used cDNA and EST data to search for over-represented motifs in 14 retrogenes expressed only in adult testis. We identified two motifs, CTSWGTGCM and SACMRWGSMMWG (Table 3 and Supplementary Table 4), that occur at a significantly higher frequency in testis-specific genes than in the genome as a whole. For the first motif, the ratios are 7/14 in the testis-specific retrogene set and 13/250 in the entire genome; for the second motif, the ratios are 8/14 in the testis-specific retrogene set and 2/250 in the entire genome (P < 10−4 for both, Fisher’s exact test). We refer to these motifs as testis specific 1 and 2 (TS1 and TS2), respectively.
Two over-represented motifs, with the consensus sequences YGSMYCHTGYKGMCC and CCSCTGCYSVTYCSC, were detected, respectively, in the upstream regions of 32 and 18 of the 34 retrogenes expressed uniquely in the testis according to FlyAtlas data (Table 3 and Supplementary Table 4). We refer to these motifs as testis specific 3 and 4 (TS3 and TS4), respectively. These two motifs show significantly higher frequency in testis-specific retrogenes than in genes throughout the genome: for the first motif, the ratios are 32/34 in the testis-specific retrogene set and 0/250 in the genome; for the second motif, they are 18/34 in the testis-specific retrogene set and 0/250 in the genome (P < 10−10 for both, Fisher’s exact test). Again, 16 of 32 occurrences (50%) and two of 18 occurrences (11%) were located inside the coding region of the nearest 5′ neighboring gene or of itself. Even excluding these cases, there is overrepresentation of these two motifs in the retrogenes compared to the 250 genes randomly sampled from the genome (P < 10−10 for both, Fisher’s exact tests).
These six over-represented motifs are not over-represented in the testis-biased genes reported by Parisi et al. [21], nor do they show any particular location or strand specificity apart from being located within −1000 and +40 bp of the TSS (data not shown). Given their location, these motifs may be cis-regulatory regions that differ from the core promoter.
Analysis of the distribution of the six motifs (TA1, TA2, TS1, TS2, TS3, and TS4) reveals additional insights. After removing instances when the motifs occur within coding regions, since these could be false positives, the analysis showed that no retrogene contains all six motifs, while 10 retrogenes contain more than one motif. Specifically, five different motifs lie upstream of CG9582; three lie upstream of CG4701, CG31003 (gskt), CG14508, CG7094, and CG2830 (Hsp60B); and two lie upstream of CG4706, CG10839, CG10838 (robl22E), CG2528, and CG32089 (Vha16-2). TA1 is detected upstream in six retrogenes with multiple motifs (Supplementary Tables 3 and 4).
Motif conservation
The comparative analysis of motifs appearing in the core promoter region of retrogenes is reported in Supplementary Table 5, based on MUSCLE alignment, FootPrinter2 results, and manual inspection. For the motifs described in Table 2, MUSCLE alignment and footprinting approaches show similar conserved motifs. Supplementary Tables 6 and 7 show conservation of known motifs across different species, based on MUSCLE alignments and FootPrinter2 analysis, respectively. One example is CG9573, where both approaches show DMv4 and DRE motifs to be conserved in D. simulans and D. sechellia. In other instances, only one of the two approaches detected the motif. For example, in the gene CG8986 (TwdlB), the alignment approach detected a TATA box in addition to Inr, while in CG18290 (Act87E), only the footprinting approach detected the TATA box in D. melanogaster.
Interestingly, the GCT motif that we describe here for the first time is over-represented in retrogenes and is conserved across other species in two instances: CG9582 and CG11401 (Trxr-2). Both of these are retrogenes showing preferential expression in the adult testis, based on the cDNA and EST data. Given that these retrogenes are older than the Drosophila genus [3] and that the motifs seem to be conserved, we conclude that these GCT elements may have originated in parallel with the emergence of the retrogenes. However, despite the apparent conservation of GCT motif sequences in several Drosophila species, the origins of this motif remain unclear. Two of the seven retrogenes in which this motif is present in the upstream region are conserved in every species: CG9582 and CG11401. In the remaining five cases, the motifs lie in a region where conservation of the D. melanogaster sequence in other species is poor or absent entirely. These results therefore fail to indicate the mutations that may have led to these GCT motifs.
New motifs identified through conservation
Table 4 reports a list of novel motifs identified by FootPrinter2 software. These are 15 additional motifs identified for retrogenes. We assessed the conservation of these motifs in species containing these retrogenes by scanning the core promoter region as well as a region from −500 to +100, in order to account for varying lengths of 5′UTR. None of these motifs is shared with the parental gene (data not shown). Details of the conservation of the newly identified motifs in the different species is shown in Supplementary Table 8. Nearly all (13) of the conserved motifs are present in all the species possessing the retrogene, leading us to conclude that they were likely present at the time of retrogene insertion, or they arose shortly thereafter through a small number of sequence changes.
Table 4.
New conserved motifs detected in retrogenes using a comparative genomics approach.
| Gene name | Motif | Position in D. melanogaster |
|---|---|---|
| CG4706 | ACAAAATT | +39 |
| CG8330 | HTATTTT | −10 |
| CG8186 | CAACACT | −3 |
| GGADTTTTKCA | +25 | |
| CG7975 | CMAAWTT | +1 |
| WAGCMATM | −69 | |
| CG7542 | ATGAAGH | −30 |
| CG11401 | AGTTGGCAG | +14 |
| CG7768 | TGCAAAT | −3 |
| GYAWATA | +9 | |
| CG9436 | GAACWGA | +15 |
| CG5650 | ARATGGCGKS | −96 |
| CG32669 | TKTATTT | −28 |
| TYATCGM | −22 | |
| CG8629 | TAATTAAATT | −60 |
Validation of the identified motifs
We checked whether any of the newly identified motifs matched known transcription factor binding sites. First, we counted the number of matched genes containing at least one of the 10 core promoter motifs reported by Ohler et al. [10]. There were 47 of 91 showing such matches (see also Supplementary Table 5). Next, we looked for matches to known transcription factor binding sites using the approach of Down et al. [22]. Using FootPrinter2, we searched for our newly identified motifs in all genes of the FlyReg database incorporating the Drosophila DNase I Footprint Database v2.0 (http://www.bioinf.manchester.ac.uk/bergman/data/Bergman2004/v2.0/Target.html). We wished to see whether any of our motifs were protected in published footprinting experiments. We conducted two comparisons. First, searched the genes for our motifs, and we did not find any retrogene in our data set with a binding site matching that for any reported transcription factor derived from crude or purified nuclear extracts [23].
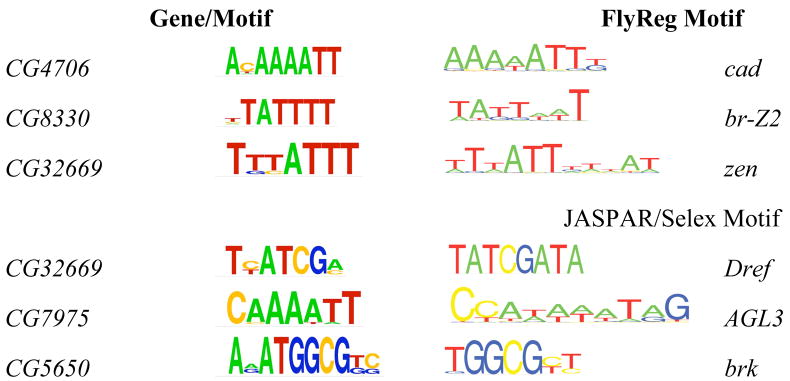
In the second comparison, we visually compared the logo/matrix of all newly identified motifs using FootPrinter2 software [24] with the FlyReg database of DNase I footprints [23]. We wished to ask whether we could assign any of our motifs to a known transcription factor. We identified three matches to FlyReg: CG4706, CG8330 (tomboy40), and CG32669 (Fig. 1). Based on the cDNA and EST data, CG4706 and CG8330 have adult testis (AT)-specific expression, and CG32669 is expressed in the larvae and pupae (LP). These motifs may contribute to the tissue-specific expression of these retrogenes.
Figure 1.
Newly discovered motifs simultaneously detected by FootPrinter2, Drosophila DNase I footprint data, and extended JASPAR CORE analysis.
Taking the three transcription factors that match the inferred motifs, we checked their associated expression information in FlyBase (http://www.flybase.org), including expressed tissue(s) and stage(s). The associated embryonic expression for cad (or caudal) is in the Malpighian tubule main body primordium; in addition, cad protein has been reported to be associated with the interphase nuclei of pole cells [22; 25]. It has been shown that br-Z2 is one of four zinc finger protein isoforms (Z1, Z2, Z3, and Z4) encoded by br (or broad). The protein product of br has been found in the salivary gland in the larval stage of flies [26]. Zen (or zerknullt) has been shown to be important in maternal regulation that controls differentiation of dorsal ectoderm in Drosophila during the embryonic stage (3–11) [27]. However, this information does not explain the expression observed for the retrogenes.
Again following the Down et al. approach [22], we conducted visual comparisons between the logo/matrix of these newly identified motifs and the extended JASPAR CORE collection. We found three matches for retrogenes: CG32669, expressed in the LP EST/cDNA library; CG7975 in the AT, EP, and EK EST/cDNA libraries; and CG5650 (Pp1-87B), expressed in the LD, RH, RE, GH, AT, SD, LP, EK, GM, EC, EN EST/cDNA libraries (Figure 1).
After checking the FlyBase for their expression pattern, we found that the protein product of Dref (or DNA replication-related element factor) has been found in the nucleus during both embryonic and adult stages. The Dref protein is a trans-activating factor that binds to the DRE of genes involved in DNA replication. Dref plays an important role in organizing zygotic expression of genes involved in DNA replication [28]. The protein product of brk (or brinker) was found in both the anterior and posterior of the dorsal mesothoracic disc in the third instar larval stage [29]. However, AGL3 was found to be a factor in Arabidopsis thaliana, based on the extended JASPAR CORE collection database. It therefore remains unclear whether flies have factors functionally equivalent to AGL3. It seems likely that flies have such a functional homologue, since this motif was also detected in a recent genome-wide analysis of promoter motifs in D. melanogaster [22]. Importantly, some of the expression results observed with the above genes may be explained by the activity of the above transcription factors.
DISCUSSION
Our results reveal an abundance of DRE, Inr, and motif 1 in parental genes, and an abundance of DRE and Inr in retrogenes. These results are consistent with the abundance of these elements revealed in previous genome-wide analyses [10; 11]. In the present study, we have identified an over-represented motif in the core promoter region of our retrogene set, which we name the GCT motif (Table 3). We infer that the GCT motif co-emerged with some of the retrogenes in our study. However, the available data do not allow us to infer what substitutions were needed in order to give rise to a particular motif. Fortuitously, our study revealed four additional new motifs in the subset of retrogenes showing adult testis expression. These motifs are found either associated with testis expression (motifs TA1 and TA2), or in testis-specific retrogenes (TS1–TS4). None of these motifs is over-represented in the testis-biased genes reported by Parisi et al. [21], indicating that they may be specific to retrogenes. There are fifteen additional motifs identified using comparative genomics approach for retrogenes. Detailed experimental analyses of all these motifs are necessary in order to determine whether they are functional regulatory motifs.
Two main questions are addressed by characterizing the promoter regions of new processed genes. First, how are regulatory sequences recruited from the target site of insertion? This report is a first attempt at understanding this process. Second and more specifically, in Drosophila, the X chromosome is inactivated in the male germline [30; 31]. It has been proposed that the genes on the inactivated chromosome move to a different chromosome to acquire testis-specific meiotic expression and to compensate for the inactivation of the parental gene [4]. This effect contributes to the demasculinization of the X chromosome in Drosophila [32]. Therefore, it will be critical for us to experimentally verify whether expression is occurring during spermatogenesis and if so, at which particular stages.
MATERIALS AND METHODS
Promoter prediction
The McPromoter package [13] was used for promoter prediction in retrogenes and parental genes. It was installed locally to detect the likely transcription start site (TSS) of these genes. We considered that the McPromoter prediction to be reliable if the hit score was higher than 0.7 [10]. We retrieved the sequences flanking putative TSSs from the D. melanogaster gene dataset (BDGP4.1) downloaded from Ensembl Multi MartView [12]. Each gene used in this analysis was classified into two classes, based on whether the 5′ end of its Ensembl annotated transcript included any nucleotides beyond the protein-coding sequence (CDS). We called a gene “annotated” if at least one of its transcripts was more than one nucleotide longer at the 5′ end than its CDS, in other words, if its 5′UTR is annotated in Ensembl. These 5′UTRs are based on EST and cDNAs, but not always on cap-trapped cDNAs [12]. Otherwise, a gene was classified as “non-annotated”. For those genes with annotated 5′ UTRs, we ran McPromoter prediction trials using a sequence extending from 500 nt upstream to 100 nt downstream of the TSS.
Motif analyses in the putative core promoter region
To find over-represented sequence motifs in the putative promoter regions of retrogenes and parental genes, we used Consensus [19], MEME [18], and visual inspection. We looked for motifs in a region between −100 and +40 relative to the TSS for genes with annotated 5′ UTRs. For those retrogenes predicted by McPromoter, the screened core promoter region was also retrieved based on the beginning and ending position (window) of the McPromoter prediction, rather than on a single reported TSS. To run the Consensus software, the following parameters were used: four (the default value) of the top matrices from each cycle were printed, and a pattern width of 8. MEME was used with a window size of 6–8 nt and the assumption that zero or one occurrence for any motif in the core promoter region was possible. MEME was asked to report the top ten most significant motifs.
Patser [15] was also used; this program scores the subsequences against supplied weight matrices. The 10 motif weight matrices reported by [10] were run through Patser using the default running parameters for the scoring process. Only the hits with positive scores were printed and parsed to analyze the motifs detected. The possible occurrences of motifs detected in each gene were positioned relative to the annotated TSS in FlyBase.
Identification of motifs in the subset of retrogenes expressed in the adult testis
MEME was run on both strands of the flanking sequence over a region extending from 1,000 nt upstream of the TSS to 40 nt downstream, including the 5′ UTR and any introns within it. This was performed for the 48 retrogenes expressed in the testis and 14 testis-specific retrogenes to discover any over-represented motifs. This enabled us to detect downstream elements if they were close to the TSS. The search window size for the possible motif was set between 5 and 50 nt, and any number of repetitions were assumed to be possible. The top ten motifs were generated, and their significance was analyzed. We used MEME to conduct a similar analysis for 34 retrogenes uniquely expressed in the testis, based on FlyAtlas data [20].
Retrieving flanking sequences for orthologs of D. melanogaster genes
The flanking sequence for an ortholog to a D. melanogaster species was considered suitable for analysis if its affiliated gene was conserved in that species. The conservation pattern for a particular D. melanogaster retrogene was examined in 11 other Drosophila species using local tBLASTn [33], with results under clear synteny conservation. The length of the 5′ UTR, including any introns, was assumed to be identical among the Drosophila species. If there was no tBLASTn hit assignment for the target sequence, or if the true upstream sequence in the contig/chromosome arm was shorter than the length intended for retrieved upstream sequence, then the flanking sequence for that gene was not extracted. The coordinates used to retrieve the flanking sequence are shown in Supplementary Table 9.
Motif conservation among species
Two approaches, alignment approach and phylogenetic footprinting, were used to search for new motifs and to reveal whether motifs identified using the previously described approaches were conserved in other species.
First, MUSCLE alignment software [34] was used to inspect the conservation pattern for the motifs reported in D. melanogaster in the region between −100 and +40 nt. For this approach we visually inspected the multiple sequence alignment results to reveal whether the D. melanogaster regions or motifs were conserved in other species.
The second approach used the motif detection software FootPrinter2 [24], which takes into consideration the phylogenetic relationship between input sequences. FootPrinter2 predicts motifs appearing in many, but not necessarily all, input orthologous sequences by constructing a phylogenetic tree for the species under study. The input phylogenetic tree of the 12 Drosophila species used in the test was (((((((simulans,sechellia),melanogaster),(yakuba,erecta)),ananassae),(pseudoobscura, persimilis)),willistoni),((mojavensis,virilis),grimshawi)).
We ran FootPrinter2 for all genes containing at least one of our 12 reported motifs. We searched between −100 and +40 nt in those regions that had clear synteny conservation based on the tBLASTn results [3]. Searched motif length was based on the particular motif consensus sequence since nucleotides beyond the consensus region are highly variable. The motif length was eight nt for DMv5, DMv4, DMv3, DMv2, and DRE; and seven for TATA-box, INR1, INR, E-box, DPE, MTE, and GCT.
To determine the consensus sequence pattern for novel motifs that might be longer than 8 nt, we combined several short overlapping motifs, and highlighted the conserved nucleotides with different colors. Therefore, the assignment was based on the longest covering region of short, overlapping motifs.
For some motifs showing less position variation, such as the TATA box, we also restricted the search to a subregion size of 50 nt, which is approximately one-tenth the recommended length of search sequence for FootPrinter2. The TATA box, Inr1, Inr, E-box, DPE, and MTE meet this criterion. The default value of 2 for the maximum parsimony score was used in FootPrinter2 during the search process. The parsimony score for any reported motif was always within this threshold. In order to qualify as novel in a given gene, the motif had to be present in the majority of species available; in the case of older genes, the motif had to be present in at least one species other than D. yakuba and D. erecta, and in the case of younger genes, it could not be absent from more than one species within the melanogaster subgroup. A similar criterion was used in the alignment approach.
To account for the fact that the 5′ UTR may have different lengths in different species and that the motif position in other species may vary, we held the search parameters constant when scanning the genes of difference species (region from −500 to +100) in order to assess their conservation of the D. melanogaster sequence. However, we looked for phylogenetic footprinting evidence of unrecognized motifs only in the core promoter region from −100 to +40. In addition, to increase accuracy, we inspected these DNA sequences to determine whether the newly identified motifs were located in the coding regions of flanking genes.
Supplementary Material
Supplementary Table 1. Features of retrogene and parental gene pairs: presence of 5′ UTR, TSS prediction by McPromoter, distribution of detected promoter motifs, and cDNA/EST hits.
Supplementary Table 2. Characteristics of the GCT motif: gene name in D. melanogaster, distance of the motif from the TSS, and putative conservation of the GCT motif in orthologous genes from other Drosophila species.
Supplementary Table 3. Characteristics of the two testis-associated, over-represented motifs TA1 and TA2 identified by MEME software for 48 retrogenes showing expression in adult testis: name of the associated retrogene, strand orientation, start position, P value as calculated by MEME, and expression information for the retrogene. The start position is reported in bold when the motif is part of a highly conserved region as annotated in the UCSC Genome Browser. CDS: motif hits within a coding region.
Supplementary Table 4. Characteristics of the four testis-specific motifs TS1 and TS2, which are based on EST/cDNA libraries, and TS3 and TS4, which are based on FlyAtlas data: name of the associated retrogenes, strand orientation, and start position. The start position is reported in bold when the motif is part of a highly conserved region as annotated in the UCSC Genome Browser. CDS: motif hits within a coding region.
Supplementary Table 5. Motif conservation information for retrogenes based on alignment and phylogenetic approaches and D. melanogaster cDNA/ESTs library hits.
Supplementary Table 6. Genes containing known motifs as revealed by MUSCLE software, their positions in D. melanogaster, and their conservation in other Drosophila species.
Supplementary Table 7. Genes containing known motifs as revealed by FootPrinter2 software, their positions in D. melanogaster, and their conservation in other Drosophila species.
Supplementary Table 8. Genes containing motifs newly identified by FootPrinter2, their positions in D. melanogaster, and their conservation in Drosophila species. Some sequences in certain genes showed additional highlighted motifs, either because additional highlighted motifs are already known (see Supplementary Tables 7 and 8), or because they were located inside the coding region of the gene or another gene. In such cases, we did not report these motifs as novel.
Supplementary Table 9. Presence or absence of retrogenes and the coordinates of the orthologous genes from 12 Drosophila species. These coordinates were used to retrieve flanking sequences for the different species.
Supplementary Figure 1. Distribution of the CA dinucleotide in the region from −100 to +40 and distribution of mononucleotides for 83 parental genes (a, c) and 65 retrogenes (b, d) possessing annotated 5′ UTRs.
Acknowledgments
We thank Uwe Ohler for providing us with the unpublished McPromoter (version 3). We are grateful to Uwe Ohler, Jiajian Liu, and Guoyan Zhao for assistance with motif detection. Research was supported by start-up funding from the University of Texas at Arlington and by grant R01 GM071813 from the National Institutes of Health (both to EB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brosius J. Retroposons--seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 2.Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–14. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Casola C, Feschotte C, Betran E. Comparative Genomics Reveals a Constant Rate of Origination and Convergent Acquisition of Functional Retrogenes in Drosophila. Genome Biology. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betrán E, Thornton K, Long M. Retroposed New Genes Out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–5. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y, Casola C, Betrán E. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics. 2008;9:241. doi: 10.1186/1471-2164-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson JJ, Kaessmann H, Betran E, Long M. Extensive Gene Traffic on the Mammalian X Chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 8.Fontanillas P, Hartl DL, Reuter M. Genome organization and gene expression shape the transposable element distribution in the Drosophila melanogaster euchromatin. PLOS Genet. 2007;3:2256–2267. doi: 10.1371/journal.pgen.0030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadonaga JT. The DPE, a core promoter element for transcription by RNA polymerase II. Exp Mol Med. 2002;34:259–64. doi: 10.1038/emm.2002.36. [DOI] [PubMed] [Google Scholar]
- 10.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohler U, Niemann H, Liao G-c, Rubin GM. Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics. 2001;17:S199–206. doi: 10.1093/bioinformatics/17.suppl_1.s199. [DOI] [PubMed] [Google Scholar]
- 14.Michiels F, Gasch A, Kaltschmidt B, Renkawitz-Pohl R. A 14 bp promoter element directs the testis specificity of the Drosophila beta 2 tubulin gene. Embo J. 1989;8:1559–65. doi: 10.1002/j.1460-2075.1989.tb03540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–77. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 16.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–45. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 17.Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–9. [PubMed] [Google Scholar]
- 19.Stormo GD, Hartzell GW., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989;86:1183–7. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 21.Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu J, Doctolero M, Vainer M, Chan C, Malley J, Eastman S, Oliver B. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Down TA, Bergman CM, Su J, Hubbard TJ. Large-Scale Discovery of Promoter Motifs in Drosophila melanogaster. PLoS Comput Biol. 2007;3:e7. doi: 10.1371/journal.pcbi.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–9. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 24.Blanchette M, Tompa M. FootPrinter: A program designed for phylogenetic footprinting. Nucleic Acids Res. 2003;31:3840–2. doi: 10.1093/nar/gkg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macdonald PM, Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324:537–45. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 26.Gonzy G, Pokholkova GV, Peronnet F, Mugat B, Demakova OV, Kotlikova IV, Lepesant JA, Zhimulev IF. Isolation and characterization of novel mutations of the Broad-Complex, a key regulatory gene of ecdysone induction in Drosophila melanogaster. Insect Biochem Mol Biol. 2002;32:121–32. doi: 10.1016/s0965-1748(01)00097-2. [DOI] [PubMed] [Google Scholar]
- 27.Rushlow C, Frasch M, Doyle H, Levine M. Maternal regulation of zerknullt: a homoeobox gene controlling differentiation of dorsal tissues in Drosophila. Nature. 1987;330:583–6. doi: 10.1038/330583a0. [DOI] [PubMed] [Google Scholar]
- 28.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J Biol Chem. 1996;271:3930–7. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 29.Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell. 1999;96:553–62. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- 30.Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci U S A. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider TD, Stephens RM. A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Features of retrogene and parental gene pairs: presence of 5′ UTR, TSS prediction by McPromoter, distribution of detected promoter motifs, and cDNA/EST hits.
Supplementary Table 2. Characteristics of the GCT motif: gene name in D. melanogaster, distance of the motif from the TSS, and putative conservation of the GCT motif in orthologous genes from other Drosophila species.
Supplementary Table 3. Characteristics of the two testis-associated, over-represented motifs TA1 and TA2 identified by MEME software for 48 retrogenes showing expression in adult testis: name of the associated retrogene, strand orientation, start position, P value as calculated by MEME, and expression information for the retrogene. The start position is reported in bold when the motif is part of a highly conserved region as annotated in the UCSC Genome Browser. CDS: motif hits within a coding region.
Supplementary Table 4. Characteristics of the four testis-specific motifs TS1 and TS2, which are based on EST/cDNA libraries, and TS3 and TS4, which are based on FlyAtlas data: name of the associated retrogenes, strand orientation, and start position. The start position is reported in bold when the motif is part of a highly conserved region as annotated in the UCSC Genome Browser. CDS: motif hits within a coding region.
Supplementary Table 5. Motif conservation information for retrogenes based on alignment and phylogenetic approaches and D. melanogaster cDNA/ESTs library hits.
Supplementary Table 6. Genes containing known motifs as revealed by MUSCLE software, their positions in D. melanogaster, and their conservation in other Drosophila species.
Supplementary Table 7. Genes containing known motifs as revealed by FootPrinter2 software, their positions in D. melanogaster, and their conservation in other Drosophila species.
Supplementary Table 8. Genes containing motifs newly identified by FootPrinter2, their positions in D. melanogaster, and their conservation in Drosophila species. Some sequences in certain genes showed additional highlighted motifs, either because additional highlighted motifs are already known (see Supplementary Tables 7 and 8), or because they were located inside the coding region of the gene or another gene. In such cases, we did not report these motifs as novel.
Supplementary Table 9. Presence or absence of retrogenes and the coordinates of the orthologous genes from 12 Drosophila species. These coordinates were used to retrieve flanking sequences for the different species.
Supplementary Figure 1. Distribution of the CA dinucleotide in the region from −100 to +40 and distribution of mononucleotides for 83 parental genes (a, c) and 65 retrogenes (b, d) possessing annotated 5′ UTRs.