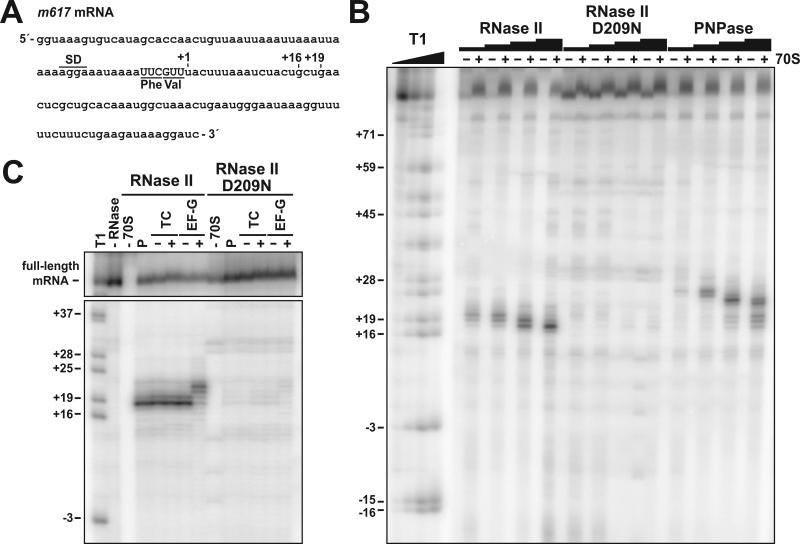
Figure 5. Exonuclease digestion of mRNA in vitro.
(A) Sequence of the bacteriophage T4 gene 32-derived m617 mRNA. The positions of the Shine-Dalgarno sequence (SD), P-site Phe codon, and A-site Val codon are indicated. Nucleotide positions are numbered with respect to the first nucleotide (+1) after the A-site codon. (B) In vitro exonuclease digestion of ribosome-bound mRNA. 5′-labeled mRNA was incubated with N-acetyl-phenylalanyl-tRNAPhe (AcPhe-tRNAPhe) with (+) or without (−) 70S ribosomes. Reactions were split and incubated with purified RNase II (4, 20, 100, and 500 nM, final concentrations throughout), RNase II (D209N) (4, 20, 100, and 500 nM), or PNPase (20, 100, 500, 2500 nM) as described in Experimental Procedures. Molecular standards were generated by digesting m617 mRNA with RNase T1 (Ambion), which cuts after G residues. (C) RNase II cleaves mRNA to the 3′ border of the ribosome. Ribosomal complexes with AcPhe-tRNAPhe in the P site (P) were incubated with valyl-tRNAVal•EF-Tu•GTP ternary complexes (+TC) to form the pretranslocation complex. In control reactions lacking ternary complexes (-TC), valyl-tRNAVal was omitted. Pre-translocation complexes were then incubated with GTP in the absence or presence of EF-G to promote translocation. Aliquots were removed after each step and incubated with 500 nM of purified RNase II or RNase II (D209N), as indicated.