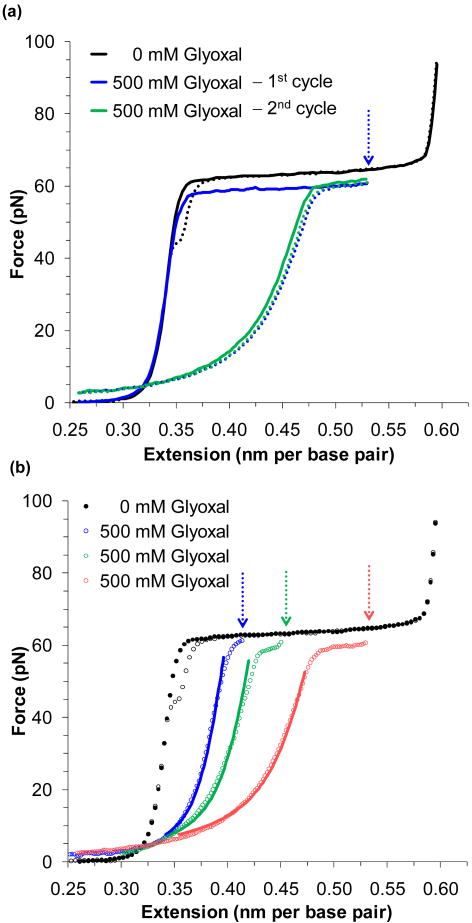
FIGURE 2.
Glyoxal binds to force melted DNA. (a) An extension/relaxation cycle of DNA is shown as a solid/dotted line in black. After stretching DNA in the presence of 500 nM of glyoxal (blue solid line), the DNA is held at the extension marked by the blue arrow for 30 minutes. Upon relaxation (blue dotted line), the strands fail to anneal, as the exposed nucleotides have been modified by glyoxal. A subsequent extension/relaxation cycle (green lines) confirms that the modification is permanent. (b) Relaxation data for DNA stretched to the lengths denoted by the arrows and corresponding to the lengths of approximately ¼, ½ and ¾ of the overstretching transition (in blue, green and red, respectively). The data is fit to a superposition of the WLC and FJC models as described in the text, for data where bss > bds. The fraction of ssDNA stabilized by glyoxal is 0.22 ± 0.03, 0.33 ± 0.01 and 0.44 ± 0.05 respectively. Figures are adapted from Shokri, et al.65