FIGURE 7.
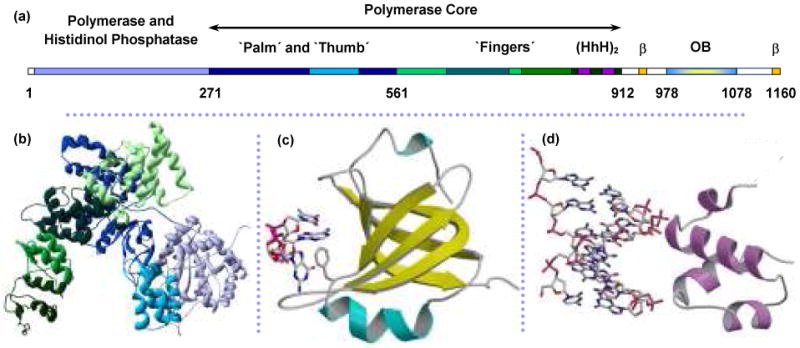
Structure of Pol III catalytic α polymerase subunit from E. coli. (a) Linear structure of α with structural domains indicated from (b) crystal structure of α amino acids 1-917.132 The polymerase core includes the palm, thumb and fingers domains that assume the classical right-hand fold. Residue numbers marking the boundaries of these regions are from the crystal structure (PDB code: 2HNH). (c) Homology model of the C-terminus predicts an OB-fold domain (not present in the crystal structure) for residues 978-1078. ssDNA is shown interacting with the residue F1031.152,162,163 (d) dsDNA is predicted to bind nonspecifically with the residues 833-889, which contains the dual helix-hairpin-helix motif, noted as (HhH)2 within the sequence map.135,136 Figures (c) and (d) are from McCauley, et al.64
