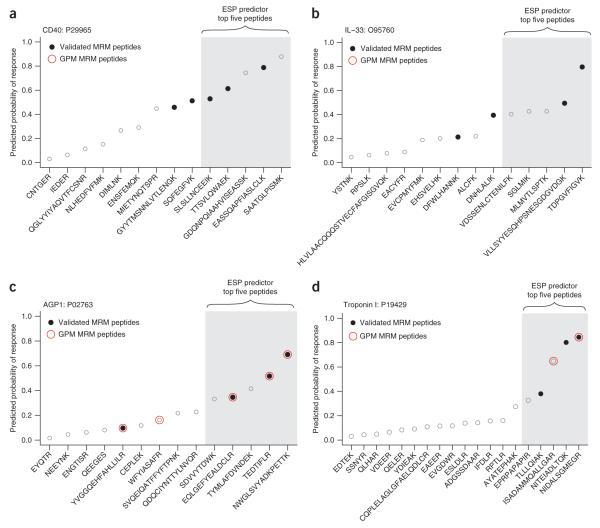
Figure 3.
ESP predictions translate into experimentally validated MRM peptides. For each protein, we performed an in silico digest (600–2,800 Da) and ensured that the top five peptides predicted by the ESP predictor were unique in the Swiss-Prot human database. Although additional filtering criteria could easily be applied after analysis with the ESP predictor, we opted for no filtering (except top five uniqueness) to demonstrate the simplicity of using the ESP predictor to select candidate signature-peptides to configure an MRM-MS assay. For all plots, peptides are sorted by the ESP predicted probability of response (y-axis). The actual rank order of measured peptide response is shown in Supplementary Table 2. (a) The ESP predictor correctly selected all three validated MRM peptides (filled black circles) out of the five predicted candidate signature-peptides for troponin I. (b) The ESP predictor correctly selected two validated MRM peptides out of the five predicted candidate signature-peptides for IL-33. In a and b, two representative proteins not found in the GPM database are shown. (c) GPM correctly selected all four of the validated MRM peptides among the top five. Three peptides are common between the ESP predictor and GPM. (d) Only two peptides were suggested by GPM of which only one was a validated MRM peptide. In c and d, two representative proteins are shown where we overlaid the MRM peptides suggested by GPM (open red circles). Example d highlights the limitations of relying solely on database predictions because two validated MRM peptides would have been missed.