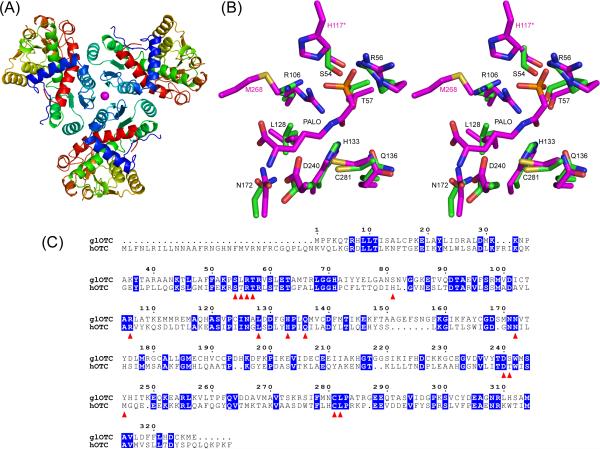
Overall fold, active site architecture of glOTC, and sequence alignment of glOTC and hOTC. (A) Ribbon diagram representation of the protein trimer. The Ni2+ ion at the center of the trimer is shown as magenta sphere. (B) Stereoscopic view of the glOTC active site superposed with the hOTC/PALO active site. The carbon atoms are colored green (glOTC) and magenta (hOTC). Other atomic colors are as follows: oxygen, red; nitrogen, blue; phosphor, orange; and sulfur, yellow. Black residue labels correspond to glOTC, except that the two residues labeled in magenta color (His117 of a neighboring subunit and Met268) are hOTC residues that are disordered in the glOTC structure (Ser82 and Tyr245, respectively). (C) Sequence alignment of glOTC and hOTC. Identical residues are blocked in blue and residues surrounding the active site are indicated by red triangles.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.