Figure 2. Functional validation of GOLPH3.
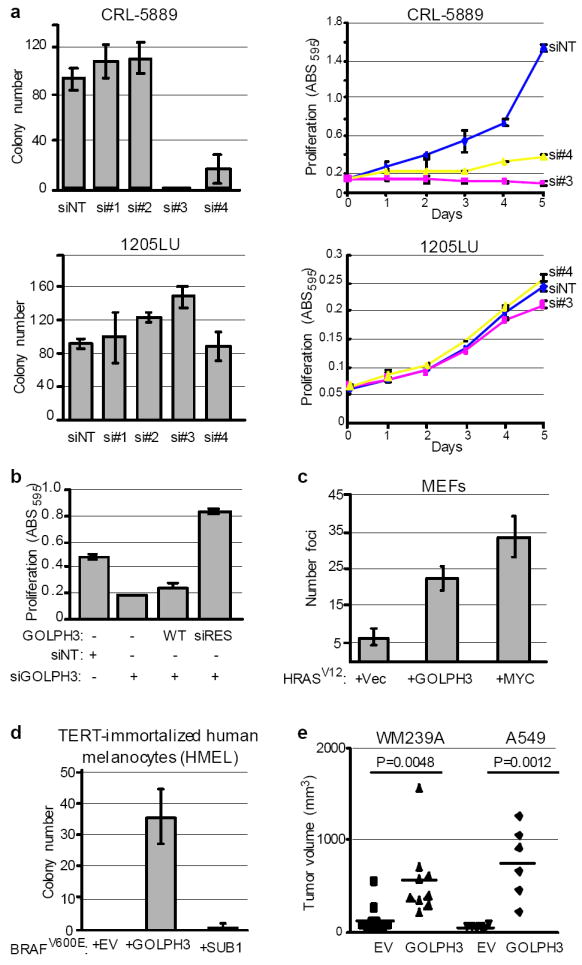
a, The indicated cell lines were treated with non-targeting siRNA (siNT) or individual siRNAs against GOLPH3 (si#1-si#4) to assay for effect on anchorage-independent growth in soft agar (left panels) and cell proliferation (right panels). Bars indicate ±S.D. b, A549 parental cells and those expressing either wild type (WT) or siRNA resistant GOLPH3 (siRES) were treated with either non-targeting siRNA (siNT) or siRNA against GOLPH3 (siGOLPH3) and assessed for effect on cell proliferation. Bars indicate ±S.D. Shown are endpoint values for day 5. c, Primary Ink4a/Arf-deficient MEFs were transfected with the indicated vectors expressing HRASV12, MYC and GOLPH3. Vec = LacZ vector control; bars indicate ±S.D.; Two-tailed t-test: HRASV12 + GOLPH3 vs. HRASV12 + Vec, p=0.0018. d, TERT-immortalized human melanocytes (HMEL) expressing activated BRAFV600E were transduced with either GOLPH3 or SUB1 to assay for effect on anchorage-independent growth in soft agar. Bars indicate ±S.D.; Two-tailed t-test for colony number: EV vs. GOLPH3, p=0.0020; EV vs. SUB1, p=0.3739. e, The indicated cell lines were transduced with GOLPH3 to assay for effect on growth of mouse xenograft tumors.
