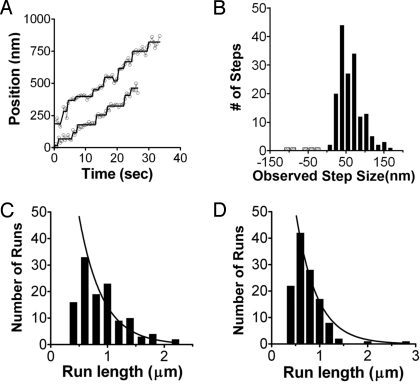
Fig. 2.
Movement of myosin VI/adaptor protein complexes on actin filaments. (A) Raw traces and steps defined by step-fitting for myosin VI/optineurin complexes. (B) Distribution of observed steps of Cy3-labeled full-length myosin VI bound to optineurin. Myosin VI molecules in which 10–20% of the molecules were labeled on the IQ-bound calmodulin with Cy3 were mixed with optineurin. The resulting complexes (two myosin VI monomers per optineurin dimer) displayed the step size distribution shown in the histogram. The average forward step size was 61.58 ± 30.67 nm (n = 163), which represents two steps of the center of mass (≈30 nm per step) of the myosin VI dimer. The average backward step size of full-length myosin VI was −20.4 ± 8.3 nm (n = 8). (C) Histogram of the distribution of the lengths of individuals for myosin VI/optineurin complexes. The average run length from pooled data for the myosin VI/optineurin complex was 0.90 ± 0.39 μm (n = 119). The distribution fit an exponential with a decay constant (λ) of 0.46 μm. (D) Histogram of the distribution of the lengths of individuals for myosin VI/tDab2 complexes. The average run length for the myosin VI/tDab2 complex (pooled data) was 0.75 ± 0.33 μm (n = 121). The distribution fit an exponential with a decay constant (λ) of 0.30 μm. For both optineurin and tDab2, the decay constants were determined by an exponential fit of the data cumulative probability distribution (not to the histogram distributions shown here), as previously described (21).