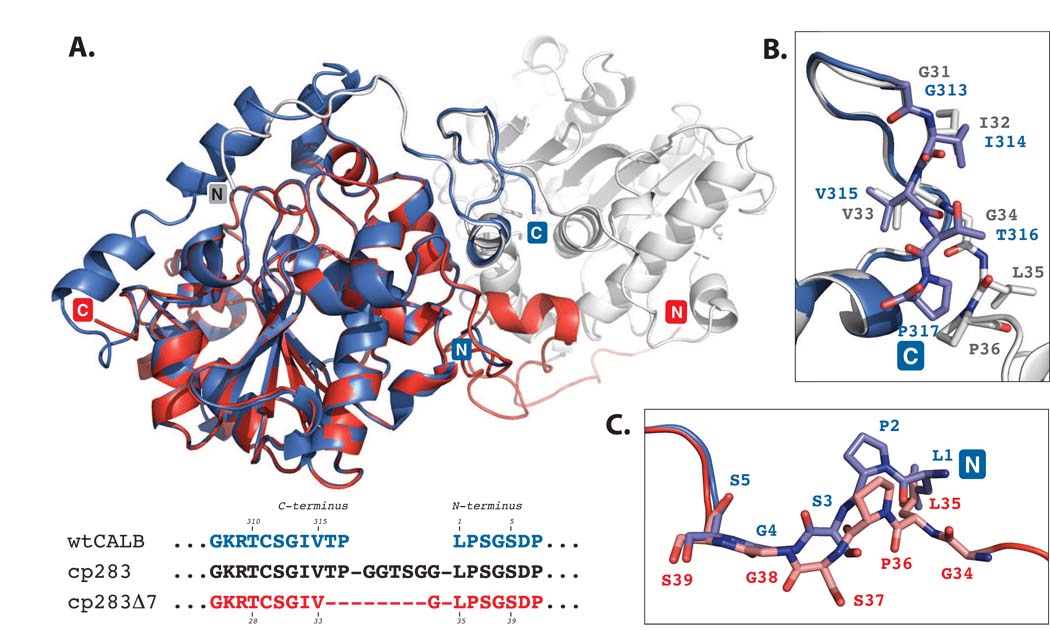
Figure 4.
Superposition of wild type CALB (PDB: 1LBS)8 and cp283Δ7 (PDB: 3ICW). CALB is colored in blue while the two subunits of cp283Δ7 are shown in red and light grey. The structure overlay is calculated based on minimal root square mean deviation (rsmd), using PyMol software. A) Overview of superimposed proteins. The core regions of the 〈/® hydrolase fold for CALB and cp283Δ7 align closely (rsmd: 0.5 Å over 263 C〈 atoms out of 298 residues). Deviations can be seen near the new termini of cp283Δ7 where the 5 N-terminal residues are slightly reoriented. The 17 amino acids at the C-terminus are invisible due to a lack of electron density, suggesting increased flexibility. B) Close-up view of the region near the C-terminus of CALB. The backbone structure and side chain orientation of the five amino acids at CALB’s carboxy end and the corresponding residues in cp283Δ7 are shown. The overlay illustrates the similar positioning of all residues up to Val315 (which corresponds to Val33 in cp283Δ7). Incremental truncation of cp283 eliminated Thr316 and Pro317, replacing them with the more flexible Gly34 from the linker sequence. Gly34 facilitates the rotation of the backbone required to properly align the domain-swapped N-terminal segment with the protein body. C) Close-up of the region at the N-terminus of CALB. A 180°-rotation at the peptide bond connecting Ser37-Gly38 (Ser3-Gly4 in CALB) results in the reorganization of Leu35-Ser37 (Leu1-Ser3 in CALB), forming a bridge to Gly34 which itself ties in with the swapped N-terminal segment of cp283Δ7.