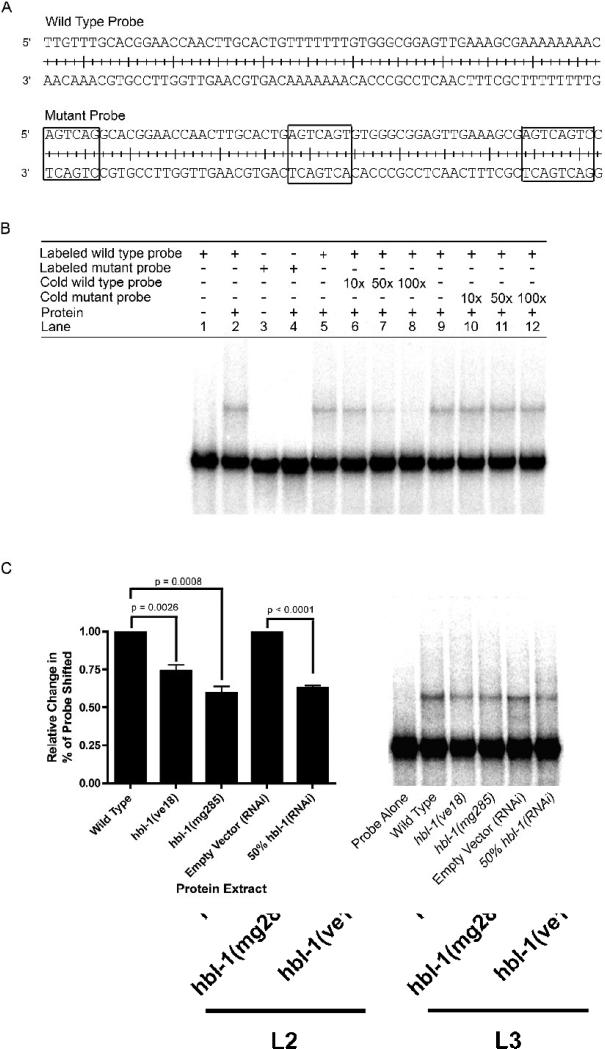
Figure 6.
A probe using the 62 bp region containing putative HBL-1 binding sites is specifically shifted in EMSAs by wild-type C. elegans protein extracts, and this shift is significantly decreased when extracts from hbl-1 loss-of-function mutants and 50% hbl-1(RNAi) are used. (A) Wild-type and mutant probe sequences used in EMSAs. Boxed sequences in the mutant probe indicate the mutations and location of potential HBL-1 sites. (B) Lanes 1−4, the wild-type probe, but not a probe with mutated HBL-1 binding sites, is shifted by C. elegans protein extract. Lanes 5−8, increasing amount of non-radiolabeled, wild-type probe (10, 50 and 100× greater than labeled probe) can compete away the labeled probe's shift. Lanes 9−12, increasing amounts of non-radiolabeled mutant probe cannot compete away wild-type probe's shift. (C) EMSAs using protein extracts from hbl-1(ve18), hbl-1(mg285) and 50% hbl-1(RNAi), have a significant decrease in the amount of probe shifted. n = 5, a representative gel is shown. Paired t-tests were used to measure statistical significance.