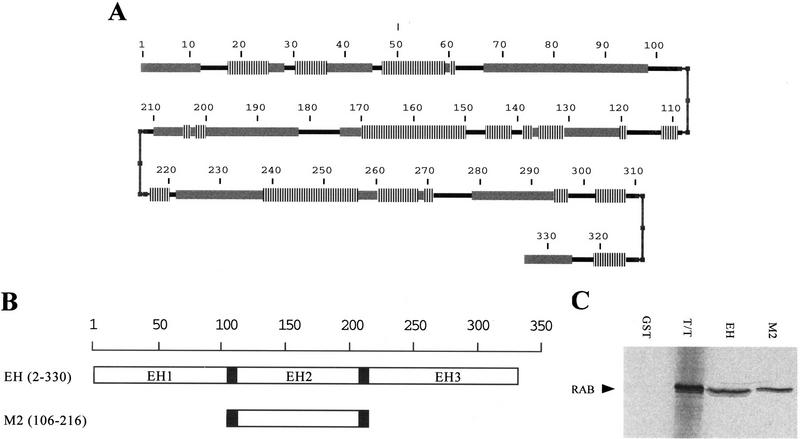
Figure 7.
Mapping of the minimal region of eps15 required for binding to NPF-containing proteins. (A) Secondary structure prediction of the amino-terminal region of Eps15 (containing the three EH domains) by a Chou–Fasman–Rose algorithm. (Vertical lines) α-Helices; (thick shaded bars) β-sheets; (thinner solid bars) coils; (small solid boxes) turn. Amino acid positions are also indicated. (B) Schematic representation of the Eps15 amino-terminal domain and of the GST fusion proteins engineered, with predicted turns indicated by solid boxes. The indicated fragments of Eps15 were engineered into GST fusion proteins and used for in vitro binding experiments. The EH construct contains all three EH domains. The M2 construct contains the region encompassing the second EH domain flanked by the natural regions pedicting the turns shown in A. Amino acid positions are also indicated in parentheses. (C) In vitro bindings. The GST fusions shown in B were used to bind the 35S-labeled RAB protein, obtained by in vitro transcription/translation of the RAB cDNA. Detection was by autoradiography. The lane marked T/T was loaded with the primary product of the in vitro transcription/translation to serve as a reference. The positions of RAB are also indicated.