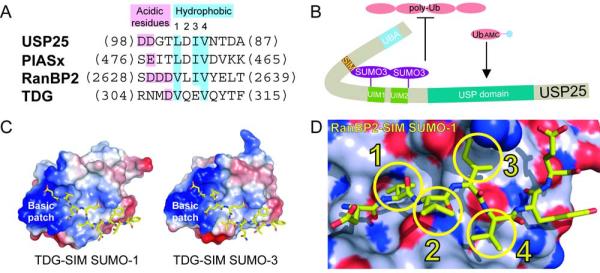
Figure 1. SUMO interacting motifs.
A) Sequence alignment for SIM elements from USP25, PIASx, RanBP2, and TDG. The latter three SIM elements are aligned based on structure. USP25 was aligned to PIASx since it exhibited the highest similarity. Acidic and hydrophobic elements are labeled and color-coded. B) A speculative model for SUMO-3-USP25 interactions wherein the SIM maintains interactions with the most N-terminal SUMO-3. This model could explain why catalytic activities are lost for polyubiquitin chains but maintained for hydrolysis of ubiquitin-AMC. C) Surface and electrostatic representations for SUMO-1 and SUMO-3 in complex with TDG SIM elements shown in yellow stick representation to highlight the basic patch (PDB 1WYW and 2D07). D) Surface representation for SUMO-1 indicating nitrogen (dark blue), oxygen (red), or carbon (light blue) in complex with the SIM from RanBP2/Nup358 colored in yellow stick representation (PDB 1Z5S). The four hydrophobic SIM amino acids are highlighted with circles and numbered 1-4 corresponding to panel A.