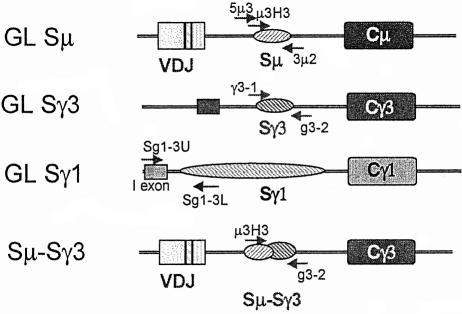
Fig. 1. Diagrams (not to scale) of the DNA segments analyzed in this study. The arrows indicate the positions of the primers used for amplification of the segments analyzed by nucleotide sequencing. See Materials and methods for the precise locations of the primers and the segments sequenced. The primer names are indicated above or below the arrows. To amplify recombined Sµ–Sγ3 segments, primers located at the 5′ end of Sµ and 3′ end of Sγ3 (µ3H3 and g3-2) were used. The 5′ primer used for GL Sµ amplification (5µ3) was located 640 nucleotides 5′ to the primer used to amplify Sµ–Sγ3 junctions. The more 5′ location of the former primer allowed us to compare our data with previously published data (Petersen et al., 2001; Nagaoka et al., 2002). The use of this primer also increased the length of the GL Sµ segment that could be sequenced from the 5′ end prior to reaching the region of Sµ that has tandemly repeated pentamers and frequently undergoes deletions. Both the 5′ and 3′ ends of the GL Sγ3 segments were sequenced to determine their mutation frequency. Both ends gave similar results, and the average is shown in Table I. Since the Sγ1 segment is too large to be amplified in its entirety, primers were designed to amplify a 2 kb segment located near the 5′ end of the Sγ1 segment, which would be deleted in cells having undergone Sµ–Sγ1 recombination at any of the sites that have been reported (Dunnick et al., 1993; W.Dunnick, personal communication). The mutation frequency was determined by sequencing the 3′ end of such fragments, which is located downstream of the Iγ1 exon, near the 5′ end of the tandem repeats. Nested primers were not used for any of the amplifications.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.