Figure 1. A current working model of the ERAD pathway in yeast.
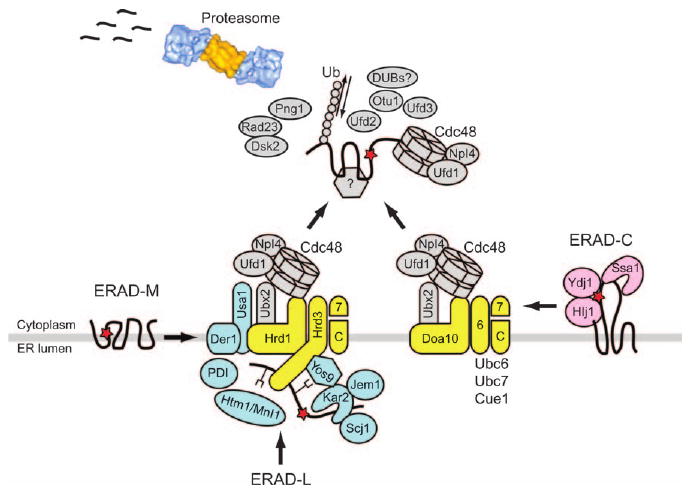
ERAD-L substrates are recognized by Hsp70/Hsp40 chaperones (Kar2, Jem1 and Scj1), PDI, putative lectins (Yos9 and Htm1/Mnl1) and the lumenal domain of Hrd3. These substrates are then retrotranslocated to the cytoplasm. In contrast, ERAD-M substrates may be directly recognized by the Hrd1 E3 ligase. ERAD-C substrates are recognized by cytoplasmic Hsp70/Hsp40 chaperones (Ssa1, Ydj1 and Hlj1) and by the Doa10 E3 ligase. The ubiquitinated substrates are delivered to the proteasome core for degradation by a series of escort factors, including the Cdc48 complex and the 19S proteasome cap. Steps following substrate ubiquitination are not yet clear for ERAD substrates. However, the polyubiquitin chain may be further remodeled by Ufd2 and by deubiquitinating enzymes (Rpn1, and/or DUBs such as Otu1). N-glycans may be cleaved by Png1, and some substrates are escorted to the proteasome by Rad23 and Dsk2. Membrane-associated E2–E3 enzymes, ERAD-L-specific components and retrotranslocation components are colored in yellow, blue and gray, respectively. The red star depicts a mutation that leads to protein misfolding, and the proteasome image was adopted from Voges et al. (126).
