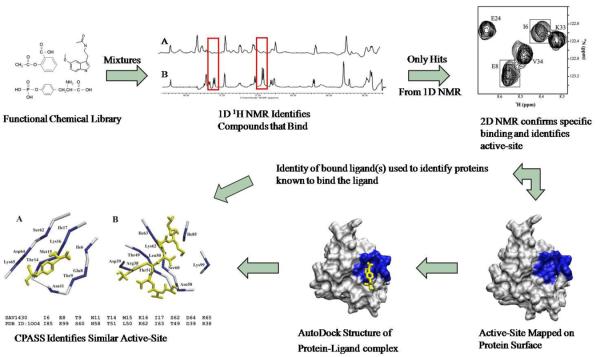
Figure 2.
Flow chart of FAST-NMR. The hypothetical proteins are screened against mixtures of ligands from the functional chemical library. Reference 1D 1H spectra of the mixtures are compared to those containing protein, where a hit is identified by changes in NMR line-width. Only the ligands identified as binding in the primary screen are further assayed in the secondary 2D 1H-15N experiment. Chemical shift changes confirm a specific interaction and identify the binding site from mapping of the CSPs on the protein's surface. The binding site and CSPs are utilized to determine a rapid co-structure using AutoDock. This co-structure is then used by CPASS to compare the ligand-defined binding site from the hypothetical protein to all other protein-ligand interactions present in the PDB. A general biological function can then be assigned based on an observed similarity to a ligand-defined binding site for a protein of known function.