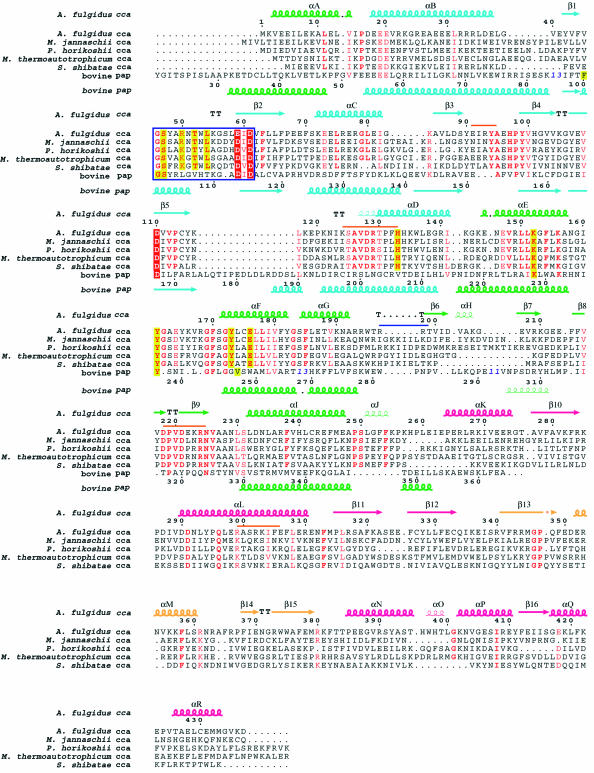
Fig. 2. Sequence alignment of the CCA-adding enzymes from five archaea (A.fulgidus, Methanococcus jannaschii, Pyrococcus horikoshii, Methanobacterium thermoautotrophicum and S.shibatae). The sequences of the N-terminal and central domains of bovine poly(A) polymerase were also aligned according to their structural composition by using O (Jones et al., 1991). Blue bold numbers indicate the number of residues not displayed. Important conserved residues are highlighted in red for catalytic carboxylates and in yellow for residues interacting with the nucleotides. Highly conserved residues are in red. The signature sequence that forms the helical-turn motif is surrounded by a blue box. The secondary structure elements of A.fulgidus CCA-adding enzyme are indicated above the alignment, and those of bovine poly(A) polymerase are indicated below the alignment, as chains (α-helices), narrow chains (310 helices), arrows (β-strands) or TT (turns). The four domains in A.fulgidus CCA-adding enzyme are color coded as in Figure 1A. The putative tRNA-binding regions are shown with an orange bar and a blue bar just above the alignment in the subunit that catalyzes the CCA-addition and the other subunit, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.