Abstract
RecA/Rad51 protein family members (Rad51, Rad51b, Rad51c, Rad51d, Xrcc2, and Xrcc3) are essential for DNA repair by homologous recombination and their role in cancers has been anticipated. Here we provide the first direct evidence for a tumor suppressor function for a member of the Rad51 family. We show that Rad51c deficiency leads to early embryonic lethality, which can be delayed on a Trp53-null background. To uncover the role of Rad51c in tumorigenesis, we have exploited the fact that Rad51c and Trp53 are both closely located on the mouse chromosome 11. We have generated double heterozygous (DH) mice carrying mutant alleles of both genes either on different (DH-trans) or on the same chromosome (DH-cis), the latter allowing for a deletion of wild-type alleles of both genes by loss of heterozygosity (LOH). DH-trans mice, in contrast to DH-cis, developed tumors with latency and spectrum similar to Trp53 heterozygous mice. Strikingly, Rad51c mutation in DH-cis mice promoted the development of tumors of specialized sebaceous glands and suppressed tumors characteristic of Trp53 mutation. In addition, DH-cis, females developed tumors significantly earlier than any other group.
Keywords: Rad51c, knockout mice, preputial gland, Rad51 paralogs, sebaceous tumor, Trp53, Muir-Torre syndrome
INTRODUCTION
DNA repair protects the genome from acquiring mutations that may potentially lead to cellular transformation and tumorigenesis. DNA double-strand breaks, the most severe type of DNA lesions, are repaired either by non-homologous end joining or by homologous recombination pathways (1). In homologous recombination, genetic information from a homologous region of a sister chromatid is used as a template to faithfully restore the damaged DNA. Rad51 is a key protein in the homologous recombination pathway, mediating strand invasion and exchange between a free DNA end proximal to a damaged site and a homologous double-stranded DNA (2). Mammalian cells possess an elaborate molecular machinery to ensure a timely and precise loading of Rad51 at sites of DNA damage. This machinery includes five members of the RecA/Rad51 family (Rad51b, Rad51c, Rad51d, Xrcc2, and Xrcc3) that show 20−30% sequence identity to Rad51 (3). These Rad51 paralogs interact with each other and can be purified as two protein complexes (4, 5). One of these complexes includes Rad51b, Rad51c, Rad51d, and Xrcc2 (BCDX2 complex), and the other contains Rad51c and Xrcc3 (CX3 complex). Functional analysis of DNA repair proficiency of double mutants in chicken DT40 cells suggests that these two protein complexes have distinct functions as Xrcc3 and Rad51d double-mutant cells display an additive effect on sensitivity to cisplatin compared with single mutants (6). Each paralog within the BCDX2 complex also appears to contribute differently to the common function based on differential sensitivity to DNA-damaging agents. Rad51c is part of both of these complexes and is thought to play a central role in these associations. Functional analysis has shown that all these genes specifically affect the homologous recombination pathway and suppress recruitment of Rad51 to the site of DNA damage (6, 7). Cell lines lacking any of these genes are sensitive to DNA crosslinking agents and are genomically unstable and accumulate chromosomal rearrangements (6, 8). In addition to these five paralogs, another member of this family, DMC1, shows 50% sequence identity to Rad51. It is a structural and functional homolog of Rad51 that functions specifically in meiotic recombination (9).
Although a link between Rad51 family members and cancer is expected, results implicating these genes in cancer thus far have been circumstantial (see ref. 10 for review and references). Overexpression of a dominant-negative form of Rad51 in CHO cells increased tumorigenesis when these cells were transplanted into nude mice. Downregulation of RAD51 was found in patients with multiple myeloma. The product of a balanced translocation between RAD51B (RAD51L1) and a high mobility group protein HMGA2, with subsequent loss of the second RAD51B allele has been implicated in uterine leiomyomas. RAD51D was found to play an important role in telomere maintenance and its E233G variant may be a low-penetrance allele in high-risk breast cancer families without mutations in BRCA1 and BRCA2. A marginal increase in risk ratio has also been found for several XRCC2 and XRCC3 sequence variants in connection with breast and some other types of cancer. RAD51C (RAD51L2) is part of a 17q23 cytoband frequently amplified in sporadic breast cancers (10, 11).
Although the above mentioned studies show association of RAD51 family members with cancer, additional verification with larger number of patients is needed (12). In addition, animal studies implicating Rad51 and its paralogs in cancer have been hampered due to early embryonic lethality of mice null for most of these genes (13-18). We have previously reported generation of a conditional allele of Rad51c in mouse, where exons 2 and 3 were floxed by insertion of a floxed PGK-neo cassette in intron 1 and a single loxP site in intron 3 of the Rad51c gene (19, 20). We used these mice to show the role of Rad51c in meiotic recombination. Here we describe generation of a null allele of Rad51c in mice. We show that Rad51c is essential for mouse embryonic development and provide evidence that Rad51c functions as a tumor suppressor. This study will set a paradigm for the role of other Rad51 family members in tumorigenesis.
MATERIALS AND METHODS
Construction of the targeting vector
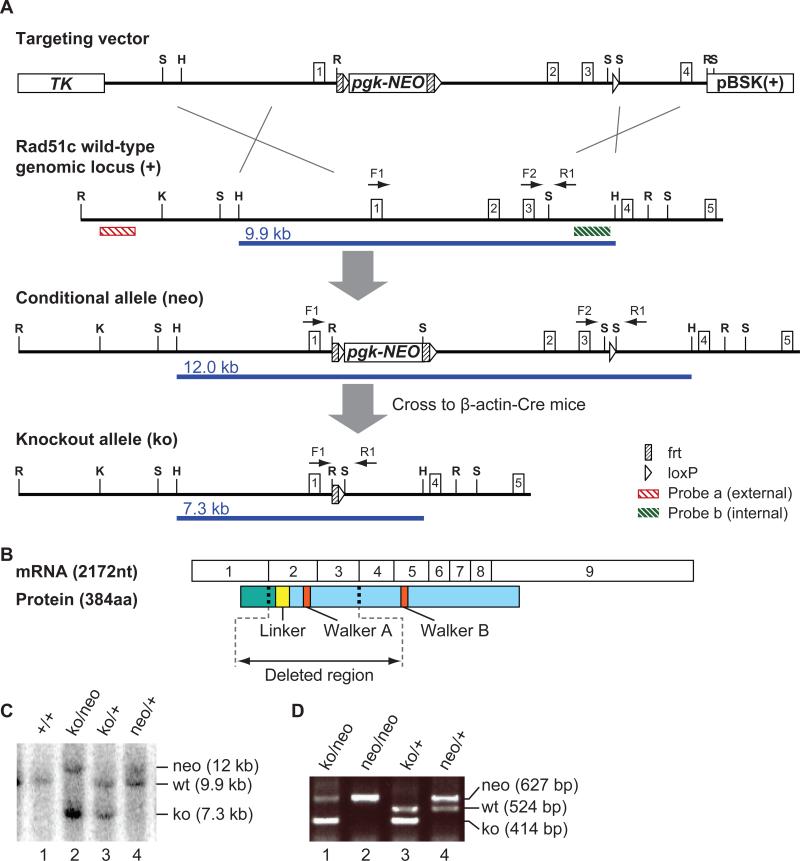
KpnI-EcoRI fragment (13.354 kb) of the genomic region containing 4 first exons of Rad51c was subcloned from the BAC clone RPCI22−514−2C into pBSK(+) plasmid to produce pCSL5 construct. Thymidine Kinase (TK) gene (NotI-BamHI fragment) under the control of the MC1 promoter was subcloned into pBSK(+) (pSKS11), and subsequently its NotI-Asp718 fragment was inserted into pCSL5 to produce pCSL5TK construct. EcoRV-BstXI fragment (2 kb) from pLMJ237 plasmid containing loxP-PGKTn5-neomycin-bp(A)-loxP was inserted into intron 3 of Rad51c in pCSL5TK construct using the lambda-Red recombineering approach (21) and the selection cassette was then deleted by Cre-mediated recombination in bacteria leaving behind a 103 bp-long insert containing a single loxP site (pCSL5TKA). In addition, an EcoRV-BstXI fragment (2 kb) from pLTM260 containing an frt-loxP-PGK-EM7-neomycin-bp(A)-frt-loxP cassette was targeted into the first intron of Rad51c using lambda-Red recombineering system (pCSL5TKAB8). The resultant targeting vector had a 6.7 kb-homology arm upstream of the first loxP site and a 3.8 kb-homology arm downstream of the last loxP site.
Targeting of Rad51c in ES cells and generation of mutant mice
Targeting vector was linearized with SalI and electroporated into CJ7 ES cells derived from 129S1/SvImJ mouse line. Electroporation and selection were performed with the CJ7 ES cell line as described elsewhere (22). 127 G418R, FIAUR ES cell clones were screened by Southern analysis of EcoRI digested genomic DNA using an external probe (see Fig. 1A) and two of them were found correctly targeted. The presence of the last loxP site was confirmed by hybridizing SpeI-digested genomic DNA with an internal probe (see Fig. 1A for the restriction site map). One of these ES cell clones was injected into C57BL/6 blastocysts to generate chimeras. One of these chimeras transmitted the targeted allele, Rad51tm1sks, denoted as Rad51cneo, in the germ line and Rad51cneo/+ pups were obtained. To produce a null allele, Rad51tm2sks, denoted as Rad51cko, Rad51cneo/+ mice were crossed to β-actin-Cre-deleter strain (23). Rad51cko/+ mice have a mixed genetic background inherited from C57BL/6 and 129S1/SvImJ mouse strains, with C3H and CD1 backgrounds from β-actin-Cre strain. The colony was maintained on a mixed genetic background. Littermate controls were used in all studies. Mice were maintained under limited access conditions at the National Cancer Institute (Frederick) and animal care was provided according to the procedures outlined in the Guide for the Care and Use of Laboratory Animals, under an approved animal care and use committee protocol.
Figure 1. Mouse Rad51c gene targeting.
(A) Scheme illustrating the gene targeting strategy to generate a Rad51c-null allele. Rad51c exons are indicated as boxes with corresponding numbers. Restriction sites are labeled as S for SalI, H - HpaI, R - EcoRI, and K - KpnI. F1, F2, and R1 designate location and direction of primers used for PCR-genotyping. HpaI restriction fragments detected by Southern analysis with the internal probe b are indicated as blue lines under each allele. A frt-loxP-PGK-EM7-neomycin-bp(A)-frt-loxP cassette was targeted into the first intron of Rad51c (at genomic location chr11: 87217150) and a single loxP site was inserted the third intron (at genomic location Chr11: 87214383) (B) Depiction of exon structure of the Rad51c transcript and the corresponding protein with functionally important regions. N-terminal domain is shown in green and C-terminal domain in shown in blue. (C) Genotyping by Southern blot showing four different genotypes with allele sizes labeled at the right. (D) Examples of genotyping using PCR primers shown in (A).
Genotyping of Rad51c and Trp53 mutant mice
For genotyping purposes, genomic DNA was extracted from tail biopsies or from frozen tumor tissues according to standard procedures. To genotype for Rad51c by Southern blot, 3−5 μg tail DNA was digested overnight with HpaI, size-fractionated in a 0.8% agarose gel in TAE buffer for approximately 6 hours to resolve 10 kb and 12 kb bands, and transferred onto a N+-Hybond nylon membrane (Amersham) by the alkaline transfer method. 531 bp-long Rad51c internal probe (Int2) was amplified from pCSL5TK plasmid (F: 5’-TGCCTGAATGTGTCTGCAC-3’; R: 5’-ATAGCAGGCAGCAGCATCT-3’) for 40 cycles at 94°C for 40 s, 57°C for 40s, and 72°C for 40 s. The PCR-product was then gel-purified and labeled with 32P-dCTP using the Random Primer Labeling Kit according to manufacturer's instructions. Hybridization was conducted according to standard procedures and band sizes corresponding to Rad51c alleles were interpreted as shown in Figure 1C. Genotyping for Trp53 by Southern blotting was performed as described elsewhere (24).
We also designed a PCR-genotyping strategy to genotype Rad51c and Trp53 mutant mice. For Rad51c, approximately 50 ng of tail or embryo DNA was amplified with the Platinum Taq polymerase (Invitrogen) using a three-primer strategy (F1: 5’-5’ACCGGGCAGTGGTGGCGCACGCCTTTAATCCCAGCACTTG -3’; F2: 5’-CAAAATGCTGGAATAATAGACCTGTGTCATACCCAAAGTG-3’; R1: 5’-GGGTATCCATATCACAGCCACTGTACTCTAGCTCCAGGAG-3’) at 94°C for 45 s, 65°C for 42s, and 72°C for 45 s. for 42 cycles (after initial 2 min at 94°C). PCR-products were analyzed by electrophoresis on a 1% agarose gel. The size of PCR products was 414 bp for a null (ko) allele, 524 bp for the wild-type, and 627 bp for the conditional (25) allele (Fig. 1D).
For Trp53, tail DNA was amplified with the Platinum Taq polymerase (Invitrogen) using a three-primer strategy (F1: 5’-CCTCAATAAGCTATTCTGCCAGCTG-3’ [P53ex5wtF]; F2: 5’-CGTGATATTGCTGAAGAGCTTGGC-3’ [P53neoF]; R1: 5’-CTGTCTTCCAGATACTCGGGATAC-3’ [P53ex6R]) at 94°C for 30 s, 55°C for 30s, and 72°C for 45 s. for 35 cycles (after initial 2 min at 94°C). PCR-products were analyzed by electrophoresis on a 1% agarose gel. The size of PCR products was 370 bp for a mutant (ko) allele, 320 bp for the wild-type allele.
Generation and aging of mouse cohorts
We studied 5 mouse cohorts as described below. We used Trp53 heterozygous mice carrying Trp53tm1Brd allele, denoted as Trp53ko (23). To produce the wild-type, Rad51cko/+ and Trp53ko/+ as well as double-heterozygous animals with mutant alleles located on different homologous chromosomes (DH-trans), we intercrossed Rad51cko/+ and Trp53ko/+ mice. Rad51c and Trp53 reside on mouse chromosome 11 only 10 cM apart and are, therefore, linked. To produce mice with mutant alleles on the same homologous chromosome (DH-cis), we first mated DH-trans mice with the wild-type and selected rare double-heterozygous progeny, which could be obtained only if the two mutant alleles recombined during meiosis. Such mice carrying the two mutant alleles on the same homologous chromosome were then we backcrossed to the wild-type to increase the cohort size. Mice were group-housed with food and water ad libitum and were maintained on a 12 hr light/dark cycle. Animals were monitored for 600 days. Sick mice and those with visible tumors were sacrificed and sent for pathological evaluation. One half of each tumor mass identify at necropsy was snap-frozen and stored at −80°C for further LOH analysis.
Immunostaining for MSH2
To test whether a mismatch repair protein Msh2 was specifically lost in preputial and Zymbal's gland tumors, we stained those tissues immunohistochemically. Paraffin slides were first deparaffinized with xylene and then rehydrated through 4 changes of 100% ethanol and one change of 95% ethanol. Endogenous peroxidase was blocked for 15 minutes in 0.6% H2O2 in methanol. Heat-induced epitope retrieval (HIER) with 0.01 M citrate buffer (BioGenex Laboratories, San Ramon, CA) was carried out at 100°C for 10 minutes. The slides were then rinsed with PBS for 10 minutes, blocked with goat serum (Vector Labs) for 30 minutes and incubated with primary anti-MSH2 antibody (Abcam, diluted 1:50 in PBS with 0.1% BSA) overnight at 4°C. Slides were then rinsed with PBS and incubated with the biotinylated secondary goat-anti-rabbit antibody (Vector Labs) for 30 minutes, diluted 1:100 in PBS with 10% goat serum, followed by an avidin-biotin peroxidase complex method using the Vector Elite ABC kit (Vector Labs, Burlingame, CA). Reaction was detected using DAB and counterstained with hematoxylin and mounted in Permount™ (Daigger, Vernon Hills, IL).
Generation of Rad51c-null MEFs in vitro and proliferation test
Due to an early postimplantation lethality of Rad51cko/ko embryos, no Rad51c-null mouse embryonic fibroblast cells (MEFs) could be established by a standard method. Therefore, we generated such cells by infecting MEFs that were homozygous for the conditional Rad51c allele (Rad51cneo/neo) with adenovirus expressing Cre-recombinase (AD-Cre-GFP, Viral Technology Laboratory, NCI-Frederick). Primary MEFs were isolated from E13.5 F1 embryos obtained from a Rad51cneo/+ mother backcrossed on C57BL/6 genetic background for 7 generations and a Rad51cneo/+ father backcrossed on 129/SvEv genetic background for 8 generations. Exponentially growing MEFs (P1) were trypsinized and resuspended in a small volume of the culture medium (DMEM supplemented with penicillin and streptomycin and 15% FBS) at 3×107 cells/ml. A 100 μl aliquot containing 3×106 cells was mixed with 3 μl AD-Cre-GFP suspension (3×107 viral particles) to achieve 10 MOI (Multiplicity Of Infection) ratio. The mixture was allowed to stand for 30 minutes at room temperature and then split between two 10 cm tissue culture dishes. Infection efficiency was evaluated the next day by GFP expression. Genotyping for Rad51c by Southern blot was used to confirm 100% recombination efficiency. Four days after infection, the cells were collected and seeded at 0.3×106 cells per one 6 cm dish in duplicates for a proliferation assay according to the 3T3 protocol. Cells were collected and counted every 3 days and seeded again at 0.3×106 cells per dish for 6 consecutive passages. Average increase in cell number was calculated for each passage and plotted as a cumulative growth.
Isolation of Rad51c-null MEFs from embryos
To isolate Rad51c-null MEFs, on Trp53-null background, we intercrossed a pair of DH-cis mice and dissected embryos at E10.5. At that point double-mutant embryos appeared morphologically similar to normal E9.5 embryos. Cells were explanted from embryo carcasses into 10 cm tissue culture dishes by standard techniques and incubated at 3% oxygen concentration to suppress cellular senescence (26). Initially, mutant cells grew poorly and had to be split at a very low dilution ratio for the first 5−7 passages. At approximately P10, the cell lines stabilized and grew at a similar speed as control MEFs (data not shown). Control Trp53ko/ko MEFs were isolated from E9.5 embryos essentially the same way.
Drug sensitivity test
We tested the effect of DNA-damaging compounds on mutant cell proliferation in vitro. Two independent cell lines were tested for each genotype: Rad51cko/ko; Trp53ko/ko double-null, Trp53ko/ko single-null control, and the wild type. We seeded 4,000 cells per well in gelatinized 24-well plates in duplicates for each line. Continuous drug treatment was started in 18 h at the following doses: mitomycin C (MMC) at 0, 10, 20, and 30 ng/ml, methylmethane sulfonate (MMS) at 0, 5, 10, and 15 μg/ml. Cells from one plate were trypsinized and counted using a Coulter counter and used as a control for plating efficiency and as a “before treatment” day1 reference. Two days later (3rd day after seeding) the remaining cells were counted the same way. Day1 reference numbers were subtracted from day 3 cell numbers to evaluate growth of each cell line. The resulting cell counts were expressed as percentages from the untreated wells.
Rad51 foci formation assay
We plated 40,000 cells per well in gelatinized Tissue Culture Treated Glass slides (Falcon). We irradiated slides 48 h later with 10 Gy. Six hours after irradiation, we fixed the cells with 4% paraformaldehyde for 5 min, washed them twice with PBS and permeabilized in PBS-buffered 0.1% Triton X-100 for 10 min. After two additional washes with PBS, we blocked cells in a blocking solution (1% BSA, 0.05% Triton X-100, 10% donkey serum in PBS). We performed antibody staining and imaging as described previously (27). 10−15 images with a total of at least 50 cells have been scored for Rad51 and γH2AX foci.
Chromosomal aberration test
We treated MEFs with colcemid (Invitrogen) for 1.5 h to arrest them at metaphase. The cells were then trypsinized, washed and resuspended in hypotonic solution at 37 °C (0.075M KCl) for 15 min and fixed in a methanol–acetic acid mixture (3:1 vol/vol). We stained air-dried preparations in Giemsa solution (10% Sorensen's buffer and 2% Giemsa, J.T. Baker). 200 well spread metaphases containing at least 40 chromosomes from each genotype were examined blindly for structural aberrations.
Statistical analysis
Animal survival and tumor latency data were processed using the survival / reliability function and P-values were estimated using the Wilcoxon test using the JMP 5.0.1a statistical software package. Numbers of Rad51 and γH2AX foci in MEFs were evaluated using basic statistics functions in JMP. Because γH2AX foci showed a bimodal distribution, we separated the cells into groups A and B depending on the number of observed foci and evaluated them separately. Deviation of genotype segregation from the Mendelian ratio at various developmental stages was calculated using chi test function in MS Excel (Supplementary Table 1).
RESULTS
Gene targeting and embryonic lethality of Rad51cko/ko mice
To generate a constitutive knockout allele, we crossed the mice carrying the floxed allele of Rad51c (Rad51tm1sks, denoted hereafter as Rad51cneo) that retains the neomycin resistance gene (25) to mice expressing Cre recombinase under the control of the human ß-actin promoter (28). The resulting allele, Rad51tm2sks denoted hereafter as Rad51cko, lacked exons 2 and 3 that code for a 142-amino acid region, including a linker region and an ATPase motif called Walker A (Fig. 1A, B). Walker A motif is indispensable for the function of Rad51 family proteins (19, 29). No mature Rad51c protein could be detected from this allele as has been shown previously (19, 29). We obtained Rad51c heterozygous (Rad51cko/+) mice that were viable and fertile and indistinguishable from their wild-type littermates.
When Rad51cko/+ mice were intercrossed, we did not obtain homozygous mutant (Rad51cko/ko) offspring (Supplementary Table 1), which suggested that these mice die during embryogenesis. In addition, the number of Rad51cko/+ newborn mice relative to the wild-type (239 and 156, respectively) deviated from the Mendelian 2:1 ratio and indicated that some Rad51cko/+ mice die during gestation. To determine the cause and time of lethality, we dissected embryos from a Rad51cko/+ intercross at various gestational stages between E5.5 – E10.5. At all stages with the exception of E8.5, approximately 25% of all embryos had an abnormal phenotype (Fig. 2 and Supplementary Table 1). Genotyping of E7.5 and E8.5 embryos confirmed that phenotypically abnormal embryos were indeed Rad51cko/ko. At E8.5, 39% of all embryos (n=126) were phenotypically abnormal. We found that 2 out of 18 phenotypically abnormal embryos at this stage were heterozygous for Rad51c while the remaining 16 Rad51cko/+ embryos were indistinguishable from the wild-type littermates (Supplementary Table 1). This partial embryonic lethality of heterozygous mice may account for a sub-Mendelian ratio of Rad51cko/+ relative to wild-type newborn mice. Although its exact cause is currently unknown, we speculate that this may be due to mixed genetic background (see Discussion section).
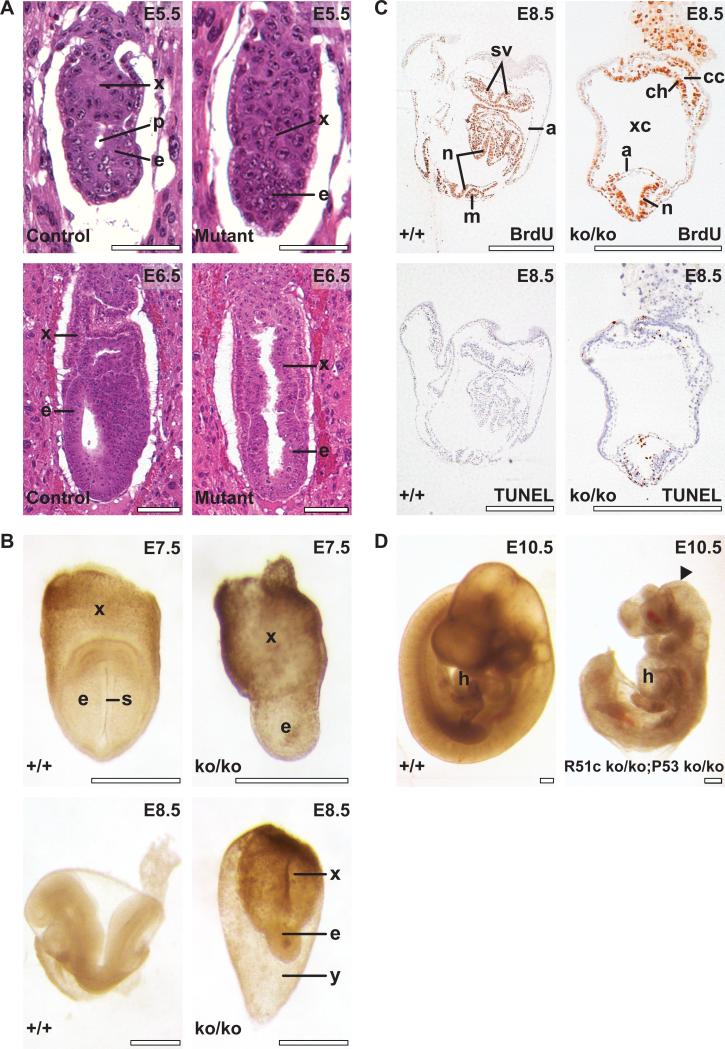
Figure 2. Rad51cko/ko mice die during early embryogenesis.
(A) Early postimplantation embryos sectioned with the deciduas and stained with hematoxylin and eosin. Top left, developmentally normal control E5.5 embrys reveals equally developed embryonic (e) and extraembryonic (x) tissues with a proamniotic cavity (p) clearly visible. Top right, mutant embryo demonstrates reduced embryonic tissues (e) without the cavity. At E6.5, development of embryonic tissues of the mutant embryo (bottom right) continues to lag behind compared with control embryos (Bottom left). (B) control (top left) and mutant (top right) embryos at E7.5. Primitive streak (s) is observed in control embryo. Morphology of control (bottom left) and mutant (bottom right) embryos at E8.5. x and e, as above; y, yolk sac. (C) Embryonic tissues in mutant embryo at E8.5 reveal active proliferation as evidenced by BrdU staining (top right) and increased apoptosis detected by TUNEL assay (bottom right) compared with control littermates (top and bottom left, respectively). n, neural folds; m, somites; a, amnion; sv, sinus venosus; xc, extraembryonic coelomic cavity; ch, prospective chorion; cc, ectoplacental cavity. Scale bar corresponds to 100 μm in A, and to 500 μm in B-D. (D) Partial rescue of the Rad51c-null phenotype on Trp53-deficient genetic background. Rad51cko/ko; Trp53ko/ko embryo at E10.5 (right) has almost normal morphology but smaller than a control Rad51cko/+; Trp53ko/+ littermate (left). Notice truncated caudal region and unclosed head folds in the mutant embryo (arrowhead). Mutant embryo shows apparently normal looking heart (h).
At E5.5 the mutant embryos lacked the proamniotic cavity and exhibited a slight delay in development (Fig. 2A, top). At E6.5 – E7.5 the delay in development of the embryo proper further increased while extraembryonic tissues continued to grow comparable to control littermates (Fig.2A, bottom and Fig. 2B top). The degeneration processes became evident after E8.5 (Fig. 2B, bottom). At this stage Rad51cko/ko embryos usually formed neural folds but looked severely abnormal and underwent resorption shortly thereafter. Histological evaluation of the Rad51cko/ko embryos did not reveal failure of any specific tissue type, but rather an overall growth defect of embryonic tissues. BrdU labeling of proliferating cells and TUNEL staining of cells with fragmented genomic DNA indicated that a marked increase in apoptosis occurred in the Rad51cko/ko embryos at the E8.5 developmental stage (Fig. 2C).
Genetic interaction between Rad51c and Trp53
Defects in DNA repair often lead to apoptosis, which can be ameliorated on a Trp53-deficient genetic background (14, 17). To examine whether the lethality of Rad51c-null embryos can be rescued by the lack of Trp53 and to have a better understanding of the cause of embryonic lethality, we crossed Rad51cko/+ mice to Trp53 heterozygous mice carrying Trp53tm1Brd allele, denoted hereafter as Trp53ko (24). We generated mice in which mutant alleles of these two genes (Rad51cko/+;Trp53ko/+) resided in the same chromosome (double heterozygous-cis, DH-cis, Fig. 3A) and, thus, were inherited as a single genetic locus. No double-homozygous mutant (Rad51cko/ko;Trp53ko/ko) offspring was obtained from DH-cis intercrosses. However, at E10.5, 25% of the embryos were confirmed to be Rad51cko/ko;Trp53ko/ko. These mutant embryos are developmentally similar to an E10.5 embryo with a normal looking heart but the embryos are smaller in size (Fig. 2D). In addition, these embryos have truncated caudal region and unclosed head folds. The fact that Rad51cko/ko embryos were severely degenerated after E8.5, and double-null embryos apparently progressed until E10.5 stage (Fig. 2B bottom and Fig. 2D), implied that embryonic development of Rad51cko/ko mice could be partially rescued by the loss of Trp53. In addition, cells from Rad51cko/ko embryos failed to proliferate in vitro but MEF lines from Rad51cko/ko;Trp53ko/ko could be successfully generated (data not shown). These results suggest that lethality of Rad51cko/ko embryos is due the apoptotic response to a DNA repair defect.
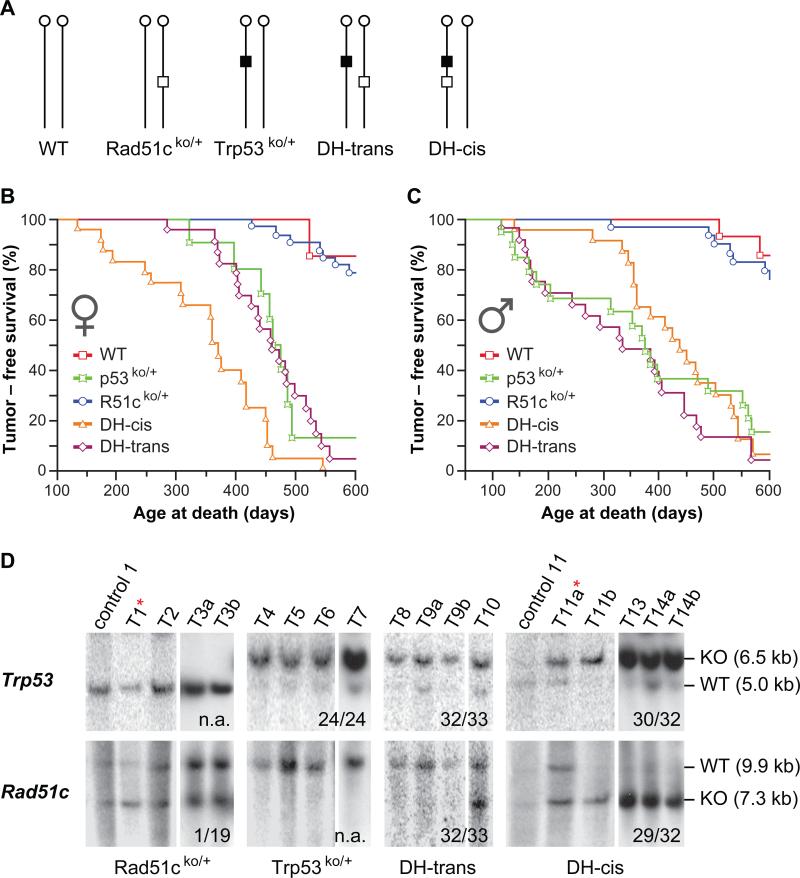
Figure 3. Interaction between Rad51c and Trp53 in mouse tumorigenesis.
(A) Schematic illustration of the five genotypic mouse cohorts used in the study. Vertical lines with a circle at the top indicate mouse chromosome 11 with a centromere. Open box specifies a mutant Rad51c allele. Closed box represents a mutant Trp53 allele. (B and C) Kaplan-Meier plot showing tumor-free survival for each group separately for females (B) and males (C). Statistical evaluation of these data is shown in Table 1. (D) Southern blot analysis of tumor tissues for LOH at Trp53 locus (upper panel) and Rad51c locus (lower panel). Animal genotype is indicated at the bottom. “T” marks tumor samples T1, sarcoma, NOS, muscle; T2, B-cell Lymphoma; T3a, Pituitary adenoma; T3b Hemangiosarcoma; T4, sarcoma, NOS, muscle; T5 rhabdomyosarcom, muscle; T6, sarcoma, NOS, muscle; T7, mammary adenoma; T8, rhabdomyosarcoma, muscle; T9a, rhabdomyosarcoma, muscle; T9b, rhabdomyosarcoma, muscle. T10, cholangiosarcoma,liver; T11a, osteosarcoma, vertebra; T11b, myoepitheliaoma, salivary gland; Letters a and b after the tumor number, indicate that the tumors are from the same animal. Control lanes show tail DNA samples for the tumor samples in the following lane. The only tumor from Rad51cko/+ animal showing LOH for Rad51c and another tumor from DH-cis animal showing LOH for Trp53 but not Rad51c is indicated with an asterisk (*) next to tumor number. Faint wild-type bands observed in LOH samples are likely be due to the presence of contaminating normal infiltrating inflammatory cells. Numbers at the bottom of each panel indicate the number of samples showing LOH relative to the total number of tumor samples tested. n.a., not applicable.
Rad51c has a gender-specific effect on tumor latency
To date there is no definitive evidence that RAD51 or any of its paralogs function as a tumor suppressor (12). It is likely that loss of any of these essential proteins triggers a severe proliferation defect, cell-cycle arrest and/or apoptosis (30). To examine the role of Rad51c in tumorigenesis, we aged Rad51cko/+ and wild-type littermates. In addition, to provide a cellular environment that lacks normal cell cycle checkpoints and allows proliferation of Rad51cko/ko cells, we also monitored the tumor predisposition of Rad51cko/+ mice on a Trp53ko/+ genetic background. We monitored the DH-cis mice (Fig. 3A), in which we expected a single loss-of-heterozygosity event to result in a simultaneous loss of the wild-type alleles of both Rad51c and Trp53. As a control, we used mice in which the mutant alleles of the two genes are on different homologues (double heterozygous-trans, DH-trans Fig. 3A). We also aged Trp53ko/+ as a second control group, to examine the effect of Trp53 loss alone.
We found no significant difference between the tumor latency of Rad51cko/+ and wild-type control littermates suggesting that heterozygosity for Rad51c alone does not increase the tumor susceptibility in mice (Table 1). However, among females, DH-cis mice developed tumors with a significantly shorter latency (369 days) than any other group including DH-trans (461 days) and Trp53ko/+ (475 days) mice (Fig. 3B and Table 1). In contrast, DH-cis males did not show a significant difference in tumor-free survival from other groups with a mutation in Trp53. Interestingly, DH-trans and Trp53ko/+ males developed tumors earlier than females of the same genotype, while the correlation was reversed for DH-cis mice. We conclude that, first, the tumor latency of DH-trans is similar to that of Trp53ko/+ mice and, second, it is functionally important whether mutations of Rad51c and Trp53 are located on the same or on different homologous chromosomes.
Table 1.
Tumor-free survival varies with sex and genotype.
| Genotype | Gender | Total number of animals | Animals with tumors | Median tumor-free survival time (days) | P value*(M vs. F) | Differences between groups* | |
|---|---|---|---|---|---|---|---|
| WT | F | 17 | 2 | > 600 | n.a. | n.a. | |
| Rad51cko/+ | F | 38 | 7 | > 600 | n.a. | vs. WT | p=0.4402 |
| Trp53ko/+ | F | 11 | 8 | 475 | n.a. | n.a. | |
| DH-trans | F | 26 | 21 | 461 | n.a. | vs. Trp53ko/+ | p=0.8635 |
| DH-cis | F | 23 | 22 | 360 | n.a. |
vs. Trp53ko/+ vs. DH-trans |
p=0.0030 p=0.0001 |
| WT | M | 21 | 3 | > 600 | 0.7839 | n.a. | |
| Rad51cko/+ | M | 32 | 6 | > 600 | 0.9765 | vs. WT | p=0.5499 |
| Trp53ko/+ | M | 20 | 16 | 385 | 0.2345 | n.a. | |
| DH-trans | M | 24 | 22 | 334 | 0.0013 | vs. Trp53ko/+ | p=0.4652 |
| DH-cis | M | 24 | 22 | 437 | 0.0466 | vs. Trp53ko/+ vs. DH-trans |
p=0.7757 p=0.0621 |
Statistically significant differences are highlighted in bold.
P-values were calculated using the Wilcoxon text.
Tumor Spectrum of DH-cis mice is different from DH-trans and Trp53ko/+
In addition to tumor latency, we also found significant differences in tumor spectrum between mice of different genotypes and genders. Consistent with previous reports (31, 32), the most common types of neoplasms in Trp53ko/+ females were osteosarcomas, mammary adenocarcinomas and lymphomas (Table 2 and Supplementary Table 2). Trp53ko/+ males, on the other hand, usually succumbed to muscle sarcomas, lung cancer, and hematopoietic malignancies. As with the tumor latency, the tumor spectrum of DH-trans mice was also remarkably similar to Trp53ko/+ mice. In contrast, DH-cis mice developed fewer tumors characteristic for Trp53-deficient mice, but revealed a greatly increased incidence of unique tumor types such as tumors of specialized sebaceous glands and tissues in the muzzle area (nasal and periocular region, Table 2).
Table 2.
DH-cis mice develop a unique tumor spectrum.
| Tumor type | Females |
Males |
||||
|---|---|---|---|---|---|---|
| Trp53ko/+ | DH-trans | DH-cis | Trp53ko/+ | DH-trans | DH-cis | |
| Osteosarcoma* | 5 (45%) | 11 (42%) | 7 (30%) | - | 1 (4%) | - |
|
Muscle sarcoma* |
1 (9%) |
- |
2 (9%) |
11 (55%) |
13 (54%) |
6 (25%) |
| Preputial gland carcinoma† | - | - | - | - | 1 (4%) | 10 (42%) |
| Zymbal's gland carcinoma† | - | 2 (8%) | - | 1 (5%) | - | 5 (21%) |
|
Muzzle area carcinomas† |
- |
1 (4%) |
4 (17%) |
2 (10%) |
2 (8%) |
5 (21%) |
| Total number of animals | 11 | 26 | 23 | 20 | 24 | 24 |
Tumor types characteristic for Trp53ko/+ mice
Tumor types characteristic for DH-cis mice
Preputial gland carcinoma was the most frequent type of sebaceous tumors in DH-cis males found in 10 out of 24 animals (42%). Four of these ten animals additionally developed Zymbal's gland carcinomas and a fifth had a Harderian gland adenoma, all being glands secreting lipids. Another animal had a Zymbal's gland carcinoma only. We also found preputial gland carcinoma in one DH-trans male and a Zymbal's gland carcinoma in two DH-trans females. In all the mice we monitored, there was only one case of Zymbal's gland carcinoma in a mouse that did not have a Rad51c mutation (in Trp53ko/+ group).
Analysis of the tumor spectrum can also explain the differences in the tumor-free survival time between genders and different genotypes. Those neoplasms that appeared to be the probable cause of death or were a primary indication for clinical sacrifice in DH-cis females included mammary gland carcinomas and carcinomas of the skin and nasal malignancies, and were the major contributors to their shorter survival (average tumor latencies of 371, 323, and 331 days, respectively). A shift from aggressive Trp53-characteristic tumor types to preputial and Zymbal's gland carcinomas and hematopoietic neoplasms in DH-cis males (463, 446, and 398 days, respectively), was probably responsible for a longer survival in this group. In Trp53ko/+ and DH-trans males, muscle sarcomas were a single tumor type that had a decisive impact on the survival time (average latency for 11 muscle sarcomas in Trp53ko/+ males was 308 days, and 255 days for 13 muscle sarcomas in DH-trans males).
LOH in tumors
In tumor cells, loss of the wild-type allele at a heterozygous locus for a tumor suppressor gene, like Trp53, is considered to be one of the primary mechanisms of tumor progression (33, 34). If LOH was indeed the leading mechanism of tumor initiation in this study, the tumor tissues from DH-trans mice would be genotypically identical to tumors from Trp53 heterozygous mice. In contrast, DH-cis mice should reveal the loss of both Rad51c and Trp53 genes. To test this, we genotyped the tumor samples collected from these animals by Southern blot analysis (Fig. 3D). As expected, all 24 tumor samples from Trp53ko/+ mice showed the loss of the wild-type allele. Similarly, with the exception of one sample, all tumors from DH-trans mice (n=33) lost the wild type copy of Trp53 and the mutant allele of Rad51c. Out of 32 tumors from DH-cis mice, 29 revealed the loss of the wild-type allele for both Trp53 and Rad51c. One DH-cis tumor sample lost a wild-type copy of Trp53 without losing Rad51c (Fig. 3D). The remaining two tumor samples did not reveal LOH for any of the two genes. Tumors from Rad51cko/+ animals (n=19) did not reveal LOH for Rad51c, except in one case. To test the possibility that Rad51c might be silenced epigenetically in these tumors, we examined its expression by RT-PCR. We found Rad51c to be expressed in all tumors tested (data not shown). Considering the fact that both the tumor latency and the spectrum in Rad51cko/+ mice were similar to the wild type mice, we concluded that Rad51c heterozygosity alone does not contribute to tumorigenesis. In contrast, when a loss of Rad51c was accompanied by a simultaneous loss of Trp53, mice developed tumors in specialized sebaceous glands.
Rad51c and sebaceous tumors
As described above, DH-cis males were highly prone to tumors of specialized sebaceous glands, particularly preputial and Zymbal's glands (Supplementary Fig. 1A and B). Sebaceous tumors from human patients often have a dysfunctional mismatch DNA repair pathway and are usually marked by downregulation of Msh2 protein expression and microsatellite instability (35). Therefore, we tested 12 sebaceous tumors for 5 microsatellite markers (D1Mit62, D15Mit93, D17Mit72, uPAR, and pro-1) and found no evidence of microsatellite instability (data not shown). We also tested three of these tumors for expression of Msh2 on paraffin sections. In all three cases the bulk of the tumor tissues stained strongly positive for this marker (Supplementary Fig. 2D), which may indicate either an upregulation of this protein in the tumors or a neoplastic transformation of already existing Msh2-positive cells normally residing at the base of each follicle (Supplementary Fig. 2B). From these results we concluded that a defect in mismatch repair was not responsible for sebaceous tumors in DH-cis mice.
In vitro phenotype of Rad51c-deficient cells explains its function as a tumor suppressor
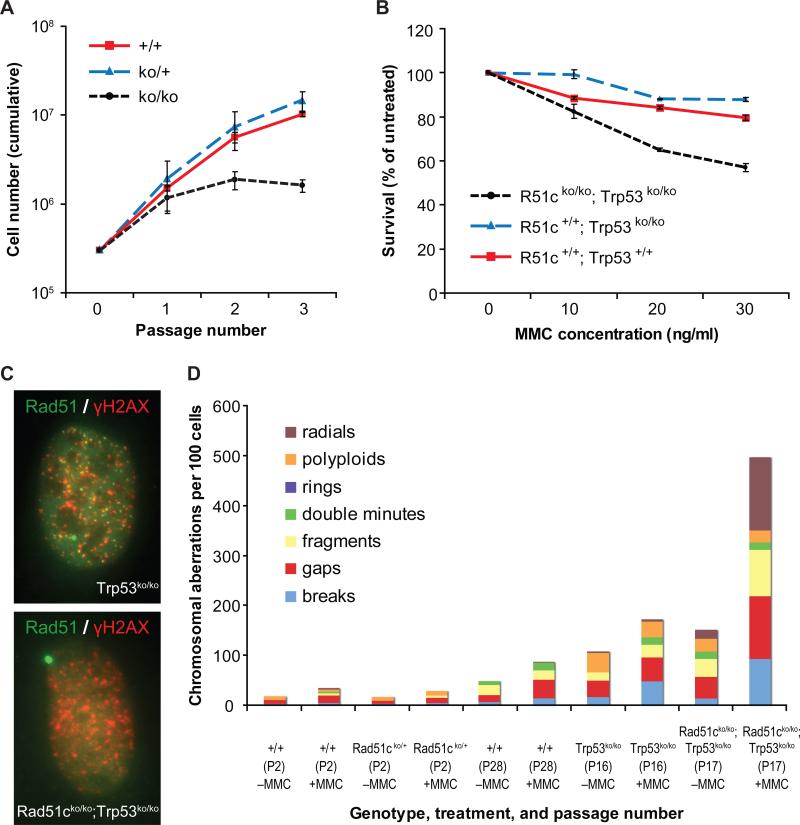
How does loss of Rad51c promote tumorigenesis? To answer this question, we investigated the effect of Rad51c loss at the cellular level. Because of the early embryonic lethality, isolation of MEFs from Rad51cko/ko embryos was not possible. Therefore, in order to generate Rad51cko/ko MEFs, we infected Rad51cneo/neo MEFs expressing a conditional allele of Rad51c with adenovirus expressing Cre recombinase. The resultant Rad51cko/ko MEFs, however, suffered a severe growth arrest after 2−3 passages (Fig. 4A), supporting the conclusion that Rad51c is essential for cell proliferation.
Figure 4. Loss of Rad51c results in proliferation defect and genomic instability in MEFs.
(A) Deletion of Rad51c induces growth arrest in primary MEFs. Wild-type, Rad51cneo/+ or Rad51cneo/neo MEFs have been infected with adenovirus carrying Cre-recombinase and proliferation has been tested using the 3T3 protocol. (B) Sensitivity of MEFs isolated from E10.5 mouse embryos to mitomycin C (MMC). (C) Rad51c-null MEFs do not form Rad51 foci 6 hours after γ-irradiation (10 Gy). Merged images are shown with Rad51 stained green and γH2AX in red. Genotype of the cell is indicated at the bottom of each image. (D) Rad51c-deficient cells accumulate various chromosomal aberrations especially after MMC treatment. The number for each aberration type is depicted proportionally in a stacked bar graph for each cell line. Genotype, passage number (P) and treatment (+ or − MMC) are indicated below each bar.
As reported previously (19), we obtained two independent Rad51c-null MEF lines from E10.5 Rad51cko/ko; Trp53ko/ko embryos (data not shown). We tested the ability of Rad51c-deficient cells to repair the DNA damage by challenging them with DNA-crosslinking agent mitomycin C (MMC) or DNA alkylating compound methylmethane sulfonate (MMS). We found the Rad51cko/ko; Trp53ko/ko MEFs to be 2−3 times more sensitive to both agents compared with Trp53ko/ko or wild-type MEFs (Fig. 4B). This observation is consistent with previously published data on Rad51c-deficient hamster CL-V4B and chicken DT40 cell lines (6, 36), although the degree of drug sensitivity of the MEFs was lower than that of hamster and chicken counterparts.
Rad51c-deficient mammalian cells have been previously reported to have attenuated Rad51 foci after ionizing irradiation, thus, implicating Rad51c in recruitment of Rad51 to sites of DNA damage and repair (6, 7). While we observed an average of 26 foci in 62% of wild-type cells (N=62) and 28 foci in 86% of Trp53ko/ko MEFs (N=55) 6 hrs after 10 Gy of γ-irradiation, no Rad51 foci could be found in Rad51cko/ko; Trp53ko/ko MEFs (Fig. 4C and Supplementary Table 3). In addition, unirradiated Rad51cko/ko; Trp53ko/ko MEFs revealed more γH2AX foci-positive cells (78% vs. 22−39% in controls) than the control MEFs suggesting the presence of abnormally large amount of damaged DNA even without exposure to any DNA damaging agent.
Increased DNA damage and dysfunctional DNA repair leads to genomic instability. We determined the frequency of various chromosomal aberrations in mutant and control MEFs with or without treatment with a low dose of MMC (10 ng/ml) corresponding to a LD10 for wild-type MEFs (Fig. 4B and C). While an average of 106 chromosomal aberrations per 100 cells was observed in untreated Trp53ko/ko MEFs, their number increased to 150 in Rad51cko/ko; Trp53ko/ko MEFs, suggesting that there is a constant level of DNA damage persisting in double-mutant cells even without a genotoxic treatment. This is consistent with the increased proportion of γH2AX-positive cells in unirradiated culture as described above. After treatment, the number of chromosomal aberrations in Rad51cko/ko; Trp53ko/ko MEFs increased to 496 per 100 cells compared with only 170 aberrations per 100 cells in Trp53ko/ko MEFs. The most frequent types of aberrations were radial structures, chromosomal fragments, chromatid gaps and breaks, which is consistent with the effect of MMC. It is worth noting that the number of chromosomal aberrations in primary wild-type and Rad51cko/+ MEFs was significantly lower compared with Rad51cko/ko; Trp53ko/ko and Trp53ko/ko MEFs . This may reflect disruption of some cellular pathways involved in chromosomal stability during establishing the cell lines in culture. Taken together, tumors arising from DH-cis mice, which became functionally null for both Rad51c and Trp53 in most cases, are predicted to be deficient in DNA repair and genetically unstable and, thus, different from DH-trans tumors. This may have had a significant influence on the tissue specificity of tumors that developed in these mice as it is known for many other DNA repair genes.
DISCUSSION
Functional analysis of Rad51 paralogs
Here we describe generation of a null allele of Rad51c and show that Rad51c is essential for viability in mice. Like Rad51c, loss of other Rad51 paralogs in mice also results in embryonic lethality. Rad51b-null embryos almost completely disappear as early as E7.5 and have the most severe phenotype among all paralogs (17). Rad51d-deficient embryos die between E9.0 and E10.0 (16). Xrcc2-mutant embryos develop normally through E8.5 (15). However, approximately 75% of the mutant embryos die between E10.5 – E12.5. Some even survive to birth but die within 20 minutes due to underinflated lungs. The phenotype associated with loss of Xrcc3 in mice is not known. Interestingly, the severity of the embryonic phenotype of Rad51b, Rad51c, and Rad51d directly correlates with respective cellular phenotypes in DT40 cells in terms of sensitivity to stalled replication forks induced by a topoisomerase I inhibitor, camptothecin, rather than relative sensitivity to DNA interstrand crosslinks induced by cisplatin (6). In spite of their overlapping functions, each paralog results in a distinct phenotype. This suggests that these paralogs may have other unique functions. Indeed, Rad51d is shown to be essential for telomere stability (37). A role in meiotic recombination and resolution of Holliday junctions (HJs) has been demonstrated for Rad51c and Xrcc3 and this function is evolutionary conserved through Arabidopsis thaliana (19, 38-40).
Rad51c is a tumor suppressor
Rad51c is evidently essential for DNA repair. Therefore, we expected an increase in tumor predisposition in Rad51cko/+ mice due to genomic instability in cells undergoing LOH at Rad51c locus. Failure to observe LOH in this group can be attributed to the fact that loss of the Rad51c function may be too detrimental for a cell, thus, causing its elimination prior to the neoplastic transformation. The fact that, during the 600-day period that we monitored the mice for tumors, the tumor latency, spectrum, and frequency did not significantly differ between Rad51cko/+ and wild-type mice, supports our conclusion that heterozygosity for Rad51c alone does not predispose mice to cancer.
Because loss of Trp53 function can delay the onset of apoptosis in cells experiencing a severe proliferation defect due to Rad51c deficiency, we examined the tumor susceptibility of Rad51cko/+ mice on a Trp53ko/+ genetic background. We monitored two classes (DH-trans and DH-cis) of genotypically identical (Rad51cko/+;Trp53ko/+) mice. It was intriguing to find that these two classes produced distinctly different tumor types. LOH analysis revealed that most of the tumors from DH-cis mice lost the wild-type alleles of both Trp53 and Rad51c while DH-trans mice lost the wild-type allele of Trp53 along with the mutant allele of Rad51c but retained its wild-type allele. This result strongly suggests that Rad51c plays an important role in tumor formation, albeit Trp53-dependent. Such dependence on Trp53 loss has been reported for many other tumor suppressor genes (e.g. Brca1, Brca2, Fbxw7/Cdc4) (41-44). Although tumor latency in DH-cis mice is primarily determined by the LOH at the Trp53 locus, additional loss of Rad51c essentially overrides the tissue-specific effect of Trp53 mutation and promotes a shift from sarcomas (mesodermal origin) to malignancies of skin and adrexia (epidermal origin) especially those of specialized sebaceous glands and other glandular and neuroepithelial tissues such as those in the nasal and periocular area. Therefore, we conclude that Rad51c should be considered a Trp53-dependent tumor suppressor. However, we cannot rule out the possibility that there are strain-specific modifiers linked to the originating alleles of the Trp53 and Rad51c knockouts that, when combined, alter the tumor spectrum in DH-cis and DH-trans mice.
Why are Rad51c-tumors tissue-specific?
Our findings raise two important questions: how does loss of Rad51c give rise to malignancies that are tissue specific in spite of its role in DNA repair in every cell, and how does addition of Rad51c-deficiency prevent development of Trp53-characteristic tumors in compound heterozygotes? In regards to the first question, examples of tissue-specific tumors caused by defects in genes that are essential for every cell type are well known. Genes like BRCA1 and BRCA2 are involved in DNA double-strand break repair but are predominantly associated with breast and ovarian cancer (45). Similarly, MLH1, PMS2, and MSH2 are important mismatch repair genes but their mutations lead to colon cancer (46, 47). The cause of the tissue-specific phenotype for these genes remains to be understood. It is possible that some downstream genes mutated in the absence of such DNA repair genes are responsible for the tissue-specific phenotype. Alternatively, these DNA repair genes may be directly involved in differentiation of certain tissues so that their dysfunction leads to a tissue-specific neoplastic transformation.
How Rad51c-deficiency can prevent development of Trp53-characteristic tumors such as to osteosarcomas and muscle sarcomas is unclear (Table 2). One possibility is that LOH for Rad51c may interfere with survival, differentiation, or neoplastic transformation of the cells that give rise to these tumors. This is supported by the observation that one of the six osteosarcoma samples from DH-cis females shows LOH only for Trp53 and not for Rad51c (Fig. 3D, see T11a). Interestingly, a myoepithelioma in the salivary gland of the same animal exhibits loss of wild-type alleles of both Rad51c and Trp53. Identification of the cell types that give rise to osteosarcomas and muscle sarcoma and the role of Rad51c in these cells will help understand the cause of reduction of these tumor types in DH-cis mice.
Tumors of specialized sebaceous glands in DH-cis mice
The high frequency of tumors in specialized sebaceous glands in DH-cis mice is unique and unexpected. Preputial gland tumors are rare in rodents (2.9% frequency reported for rats and even less for mice) (48) and have been reported to develop in only a few mouse models (49), (50). In humans, 75% of sebaceous tumors arise in the periocular region, often from a specialized sebaceous gland of the eyelid, the meibomian gland (similar to DH-cis mouse tumor shown in Supplementary Fig. 1E) (51, 52). This type of malignancy is rare but aggressive and represents 1−5.5% of eyelid malignancies (52). A small portion of sebaceous tumors is associated with Muir-Torre syndrome (53). The etiology of sebaceous cancers is unclear, but most MTS patients were found deficient in the mismatch repair pathway, primarily due to loss of MSH2 protein (54). In tumors from DH-cis mice we did not find any loss or downregulation of Msh2 or microsatellite instability, suggesting that the mismatch repair mechanism is not involved.
Partial lethality of Rad51cko/+: haploinsufficiency or a modifier effect?
Some Rad51cko/+ embryos also had an abnormal phenotype and the ratio of viable Rad51cko/+ mice was sub-Mendelian relative to the wild-type. Haploinsufficiency is unlikely to be the cause of partial Rad51cko/+ lethality because no lethality was observed previously for Rad51cko/neo mice expressing only 15−40% of the normal protein level (19, 38-40). We speculate that the presence of genetic modifiers may play a role. It is possible that one or several alleles co-segregating with the mutant allele of Rad51c may have overtly affected survival.
In conclusion, our results suggest that Rad51c functions as a tumor suppressor in mice. This is the first demonstration of a role in tumorigenesis for any Rad51 family member in mice. Similar studies may reveal unexpected tissue-specific effects for other Rad51 family members. Our future studies will be focused on examining the role of Rad51c in human tumors and on understanding the tissue-specific functions of Rad51c in epithelial tissues.
Supplementary Material
Acknowledgements
We thank J. Acharya, K. Biswas, S. Chang, I. Daar, S. Philip, K.Reilly, E. Sterneck and L. Tessarollo for helpful discussions and critical review of the manuscript. We also thank B. Martin, E. Southon, S. Reid, D. Butcher, and S. Burkett for technical assistance; J. Wada for illustrations; M. Lewandoski for providing the β-actin-Cre mouse strain. The research was sponsored by the Center for Cancer Research, National Cancer Institute, US National Institutes of Health.
REFERENCES
- 1.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–47. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 3.Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Med Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- 4.Masson JY, Tarsounas MC, Stasiak AZ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci U S A. 2001;98:8440–6. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonetani Y, Hochegger H, Sonoda E, et al. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–52. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Veelen LR, Essers J, van de Rakt MW, et al. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Takata M, Sasaki MS, Tachiiri S, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–66. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pittman DL, Cobb J, Schimenti KJ, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 10.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–35. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Bärlund M, Tirkkonen M, Forozan F, et al. Increased copy number at 17q22-q24 by CGH in breast cancer is due to high-level amplification of two separate regions. Genes Chromosomes Cancer. 1997;20:372–6. doi: 10.1002/(sici)1098-2264(199712)20:4<372::aid-gcc8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Parssinen J, Kuukasjarvi T, Karhu R, Kallioniemi A. High-level amplification at 17q23 leads to coordinated overexpression of multiple adjacent genes in breast cancer. Br J Cancer. 2007;96:1258–64. doi: 10.1038/sj.bjc.6603692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–43. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adam J, Deans B, Thacker J. A role for Xrcc2 in the early stages of mouse development. DNA Repair (Amst) 2007;6:224–34. doi: 10.1016/j.dnarep.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. Embo J. 2000;19:6675–85. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis. 2000;26:167–73. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(−/−) background. Mol Cell Biol. 1999;19:8686–93. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuzuki T, Fujii Y, Sakumi K, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93:6236–40. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznetsov S, Pellegrini M, Shuda K, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 2007;176:581–92. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leasure CS, Chandler J, Gilbert DJ, et al. Sequence, chromosomal location and expression analysis of the murine homologue of human RAD51L2/RAD51C. Gene. 2001;271:59–67. doi: 10.1016/s0378-1119(01)00498-x. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Jenkins NA, Copeland NG. A Highly Efficient Recombineering-Based Method for Generating Conditional Knockout Mutations. Genome Res. 2003;1:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessarollo L. Manipulating mouse embryonic stem cells. Methods Mol Biol. 2001;158:47–63. doi: 10.1385/1-59259-220-1:47. [DOI] [PubMed] [Google Scholar]
- 23.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–41. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 24.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 25.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 26.Parrinello S, Samper E, Krtolica A, et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsov SG, Liu P, Sharan SK. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–81. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–68. [PubMed] [Google Scholar]
- 29.French CA, Tambini CE, Thacker J. Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J Biol Chem. 2003;278:45445–50. doi: 10.1074/jbc.M308621200. [DOI] [PubMed] [Google Scholar]
- 30.Brown EJ. Analysis of cell cycle progression and genomic integrity in early lethal knockouts. Methods Mol Biol. 2004;280:201–12. doi: 10.1385/1-59259-788-2:201. [DOI] [PubMed] [Google Scholar]
- 31.Kuperwasser C, Hurlbut GD, Kittrell FS, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–9. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang SJ, Lozano G, Amos CI, Strong LC. Germline p53 mutations in a cohort with childhood sarcoma: sex differences in cancer risk. Am J Hum Genet. 2003;72:975–83. doi: 10.1086/374567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192–8. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey M, McArthur MJ, Montgomery CA, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–9. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 35.Entius MM, Keller JJ, Drillenburg P, et al. Microsatellite instability and expression of hMLH-1 and hMSH-2 in sebaceous gland carcinomas as markers for Muir-Torre syndrome. Clin Cancer Res. 2000;6:1784–9. [PubMed] [Google Scholar]
- 36.Wojcik A, Stoilov L, Szumiel I, Legerski R, Obe G. Rad51C-deficient CL-V4B cells exhibit normal levels of mitomycin C-induced SCEs but reduced levels of UVC-induced SCEs. Biochem Biophys Res Commun. 2005;326:805–10. doi: 10.1016/j.bbrc.2004.11.113. [DOI] [PubMed] [Google Scholar]
- 37.Tarsounas M, Munoz P, Claas A, et al. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–47. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 38.Bleuyard JY, Gallego ME, Savigny F, White CI. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005;41:533–45. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Masson JY, Shah R, O'Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–6. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Tarsounas M, O'Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem. 2006;282:1973–9. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Holstege H, van der Gulden H, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–6. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frappart PO, Lee Y, Lamont J, McKinnon PJ. BRCA2 is required for neurogenesis and suppression of medulloblastoma. Embo J. 2007;26:2732–42. doi: 10.1038/sj.emboj.7601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung AM, Elia A, Tsao MS, et al. Brca2 deficiency does not impair mammary epithelium development but promotes mammary adenocarcinoma formation in p53(+/−) mutant mice. Cancer Res. 2004;64:1959–65. doi: 10.1158/0008-5472.can-03-2270. [DOI] [PubMed] [Google Scholar]
- 44.Mao JH, Perez-Losada J, Wu D, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 45.Ponzone R, Baum M. The BRCA paradox in breast and ovarian cancer. Eur J Cancer. 1998;34:966–7. doi: 10.1016/s0959-8049(98)00060-4. [DOI] [PubMed] [Google Scholar]
- 46.Liu B, Parsons R, Papadopoulos N, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–74. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 47.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–38. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 48.Mitsumori K, Elwell MR. Proliferative lesions in the male reproductive system of F344 rats and B6C3F1 mice: incidence and classification. Environ Health Perspect. 1988;77:11–21. doi: 10.1289/ehp.887711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong LY, Fidanza V, Zanesi N, et al. Muir-Torre-like syndrome in Fhit-deficient mice. Proc Natl Acad Sci U S A. 2000;97:4742–7. doi: 10.1073/pnas.080063497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niemann C, Owens DM, Schettina P, Watt FM. Dual role of inactivating Lef1 mutations in epidermis: tumor promotion and specification of tumor type. Cancer Res. 2007;67:2916–21. doi: 10.1158/0008-5472.CAN-06-3427. [DOI] [PubMed] [Google Scholar]
- 51.Zurcher M, Hintschich CR, Garner A, Bunce C, Collin JR. Sebaceous carcinoma of the eyelid: a clinicopathological study. Br J Ophthalmol. 1998;82:1049–55. doi: 10.1136/bjo.82.9.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai TF, Huilgol SC, Selva D, James CL. Eyelid sebaceous carcinoma masquerading as in situ squamous cell carcinoma. Dermatol Surg. 2004;30:222–5. doi: 10.1111/j.1524-4725.2004.30069.x. [DOI] [PubMed] [Google Scholar]
- 53.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldberg M, Rummelt C, Foja S, Holbach LM, Ballhausen WG. Different genetic pathways in the development of periocular sebaceous gland carcinomas in presumptive Muir-Torre syndrome patients. Hum Mutat. 2006;27:155–62. doi: 10.1002/humu.20281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.