Abstract
We have recently shown that the anthrax lethal toxin (LeTx) selectively represses nuclear hormone receptors. In the current studies, we found that LeTx repressed activation of the MMTV promoter related to over-expression of the transcription factors, HNF3, Oct1 and c-Jun. LeTx transcriptional repression was associated with a decrease in protein levels of these transcription factors in a lethal factor (LF) protease activity dependent manner. Early administration of LeTx antagonists partially or completely abolished the repressive effects of LeTx. In contrast to the rapid cleavage of MAPKKs by LeTx, the degradation of these transcription factors occurred at a relatively late stage after LeTx treatment. In addition, LeTx repressed phorbol 12-myristate 13-acetate (PMA) -induced MMTV promoter activity and PMA-induction of endogenous c-Jun protein. Collectively, these findings suggest that transcription factors are intracellular targets of LeTx and expand our understanding of the molecular action of LeTx at a later stage of low-dose exposure.
Keywords: anthrax lethal toxin (LeTx), MMTV promoter, transcription factors, over-expression, repression
Introduction
Bacillus anthracis secretes three proteins, protective antigen (PA), lethal factor (LF) and edema factor (EF), which individually are nontoxic. One of these proteins, PA, binds to cellular receptors, is cleaved by a furin-family protease into a 63 kDa fragment that oligomerizes on the cell surface to form a heptamer. This fragment then binds and transports the two other proteins, lethal factor (LF) and edema factor (EF) into the host cell cytoplasm (reviewed in1–4). PA and EF together comprise the edema toxin; likewise, PA and LF together form anthrax lethal toxin (LeTx). LeTx is known to be the major cause of anthrax pathogenesis3; 5 and LeTx alone is sufficient to cause death when injected intravenously into laboratory animals.6
Although the endocytic transportation process is well documented, the mechanism of intracellular LF action is poorly understood. LF is a metalloprotease that cleaves and inactivates members of the mitogen activated protein kinase kinase (MAPKK/MEK) family7–12 and causes rapid lysis in macrophages of some inbred mouse strains.12 However, the fact that LeTx-resistant and -sensitive cells and mice show similar MAPKK/MEK cleavage in response to LF suggests that the cleavage of MAPKK/MEK cannot alone account for differential susceptibility or resistance to the toxin.9; 10 It is also noteworthy that the majority of LeTx studies focused on the early responses of host cells or animals within two hours after exposure to lethal or sub-lethal doses of the toxins.13–15 The molecular actions of LeTx at later stages of low-dose exposure are less well studied.
We have recently shown that LeTx selectively represses nuclear hormone receptors including the glucocorticoid receptor (GR).16; 17 We postulate that this effect might have an impact on the health of the infected individual/animal, since a fully functional hypothalamic-pituitary–adrenal (HPA) axis and resultant glucocorticoid responses are critical for survival from a number of inflammatory and infectious insults and impaired HPA axis responsiveness is associated with enhanced susceptibility to these diseases.18–25 Our studies also showed a differential effect of LeTx on nuclear hormone receptor repression when comparing a complex promoter such as the mouse mammary tumor virus (MMTV) promoter to a simple reporter driven by a glucocorticoid response element (GRE). In addition, we also demonstrated that LeTx prevents GR-DNA binding in vitro without loss of GR numbers or loss of nuclear translocation, suggesting that other accessory proteins might be targets for LeTx.16; 17
Transcriptional regulation of genes is a complicated process, which not only involves the target gene promoters, but also implicates the recruitment of intermediate factors, co-activators and proteins of the general transcriptional machinery.26–28 The long terminal repeat (LTR) of the proviral DNA of MMTV is a widely studied GR regulated promoter. Transcription of the MMTV LTR is induced by several classes of steroid hormones including glucocorticoids, progestins and androgens.29–31 The LTR of the MMTV promoter used in our previous studies also contains binding sites for other transcriptional factors that are known to activate the MMTV promoter.30; 32; 33 These include hepatocyte nuclear factor 3 (HNF3),34 the octamer-binding protein 1 (Oct1),35; 36 and activator protein 1 (AP-1) (which consists of c-Jun and c-Fos).35; 37 These transcription factors enhance the basal transcriptional activity of the MMTV promoter34; 38 and, as such, are potential targets of LeTx that might result in or enhance the transcriptional repression of the MMTV promoter described in our previous reports.16; 17 The current studies were performed to investigate whether such transcription factors are involved in the LeTx repression of MMTV transcription.
In agreement with the scientific literature, our current study shows that over-expression of HNF3, Oct1 and AP-1 increases activity of the MMTV promoter. In addition, LeTx repressed this transcription factor-enhanced MMTV promoter activity. The transcriptional repression by LeTx was associated with a decrease in protein levels of these transcription factors at relatively later stages after LeTx exposure. The proteolytic inactive LF mutant did not repress activation of the MMTV promoter by these transcription factors and had no effect on protein expression indicating that these effects of LeTx are LF protease activity dependent. These findings suggest that these transcription factors are intracellular novel targets of LeTx and expand our understanding of molecular action of LeTx at a relatively late stage of exposure.
Results
LeTx represses transcription factor enhanced basal transcriptional activity of the MMTV promoter
In order to investigate whether other transcription factors are involved in LeTx repression of the MMTV promoter, Cos7 cells were transient transfected with the full-length MMTV LTR and various transcription factors that are known to bind to this promoter. As shown in Fig. 1a, transiently transfected HNF3 increased basal MMTV promoter activity in a dose-dependent manner (open bars) and this increased transcriptional activity of the MMTV promoter was significantly repressed (30–50%) by LeTx (10 ng/ml LF + 500 ng/ml PA) (solid bars). Similarly, transiently transfected Oct1 (Fig. 1b), c-Jun (Fig. 1c), and AP-1 (c-Jun and c-Fos) (Fig. 1d), but not c-Fos alone (data not shown), also increased basal MMTV activity. LeTx significantly repressed MMTV promoter activity induced by these transcription factors. These data illustrate that LeTx represses induction of the MMTV promoter by other transcription factors in addition to nuclear hormone receptors.
Fig 1. Repression of transcription factor enhanced basal MMTV activity by LeTx.
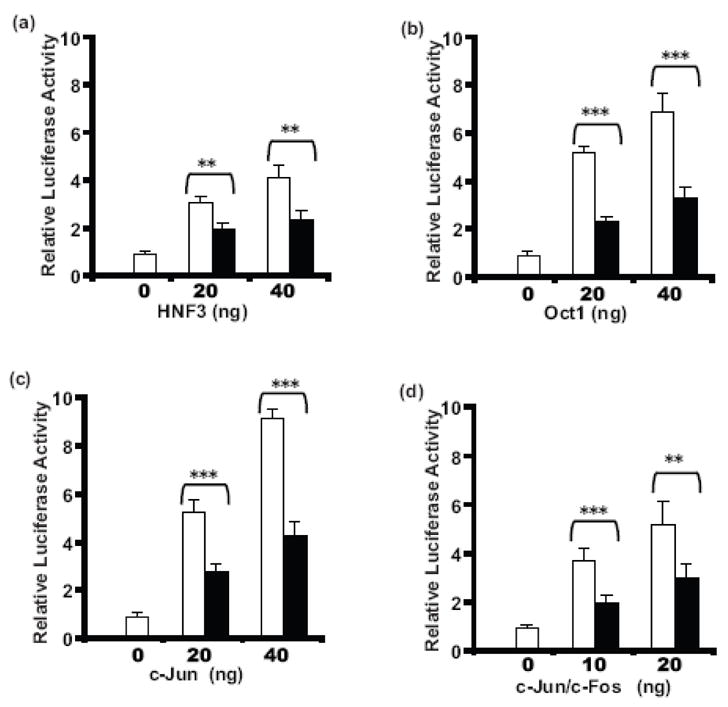
Cos7 cells plated in 24-well plates were transfected with 100 ng pGL3-LTR-luc, 15 ng phRL-TK, in the absence or presence of indicated amounts of expression plasmids for specific transcription factors using FuGENE6. After 24 h incubation in media with or without LeTx (10 ng/ml LF + 500 ng/ml PA), the cells were lysed, and luciferase assayed. The effects of 20 or 40 ng/well of the transcription factors HNF3 (a), Oct1 (b), c-Jun (c), or a combination of c-Jun and c-Fos, each at 10 ng/well or 20 ng/well (d) on the transcriptional activity of the MMTV promoter in the presence (closed bars) or absence (open bars) of LeTx are shown. Each treatment was performed in triplicates, means and standard deviations from 3 independent experiments are shown. Two-way ANOVA followed by Bonferroni post-hoc test was performed (*, p = 0.01–0.05; * *, p = 0.001–0.01; ***, p ≤0.001).
LeTx represses PMA-induced AP-1 activation of the MMTV promoter
In order to determine if LeTx is able to repress transcription factor-induced activation of the MMTV promoter as well as basal activity, the effect of LeTx on PMA activation of AP-1 (c-Jun/c-Fos) was tested in the presence or absence of LeTx (10 ng/ml LF + 500 ng/ml PA). As shown in Fig. 2, PMA induced AP-1 activation of the MMTV promoter in a dose-dependent manner (open bars). LeTx significantly repressed PMA-induced AP-1 activity (solid bars). These data indicate that LeTx represses activated transcription factor induction of the MMTV promoter, as demonstrated through PMA induction of AP-1.
Fig 2. LeTx repression of PMA-induced AP-1 activation of the MMTV promoter.
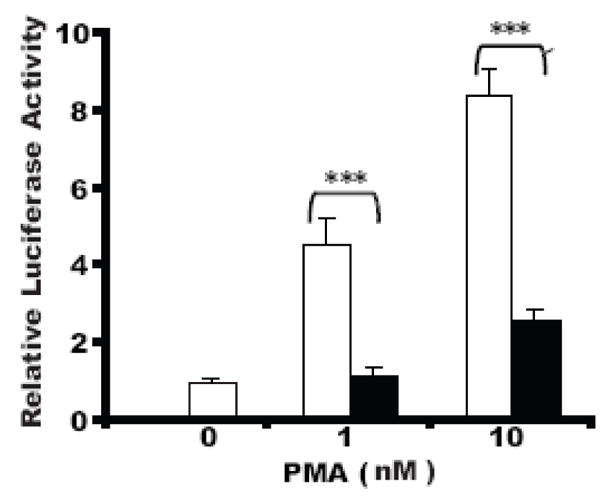
Cos7 cells plated in 24-well plates were transfected with 100 ng pGL3-LTR-luc, 15 ng phRL-TK, 10 ng c-Jun and 10 ng c-Fos expression plasmids using FuGENE6. After 24 h incubation in media with vehicle only, 1 nM or 10 nM PMA in the presence (solid bars) or absence (open bars) of LeTx (10 ng/ml LF + 500 ng/ml PA), cells were lysed, and the luciferase assayed. Each treatment was performed in triplicates, means and standard deviations of the relative luciferase activities (vehicle only was normalized to 1). 2 separate experiments are shown. A two-way ANOVA followed by Bonferroni post-hoc test was performed (*, p = 0.01–0.05; **, p = 0.001–0.01; ***, p ≤0.001).
LeTx repression of gene activity is protease dependent
The proteolytic activity of LF is vital for the lethal activity of LeTx observed in cultured cells and in animals.8 Several types of inhibitors, including small peptide substrates (e.g. IN-2-LF),7; 39–41 inhibitors of furin (e.g D9R- (D-Arg)9-NH2)40 and anti-serum against PA or LF have been found to efficiently prevent the toxicity of LeTx.42–44 To test whether the proteolytic function of LF is required for LeTx repression of transcription factor-mediated activation of the MMTV promoter, the effects of a proteolytically inactive mutant of LF (E687C) plus PA (m-LeTx)7 and inhibitors of LeTx39; 40; 43 were tested. Cos7 cells were transiently transfected with pGL3-MMTV-luc, and Oct1, c-Jun or HNF3 expression plasmids and treated with wild type LeTx (10 ng/ml LF + 500 ng/ml PA) plus various combinations of LeTx inhibitors or m-LeTx (10 ng/ml mutant LF (E687A) + 500 ng/ml PA) alone. As shown in Fig. 3a, LeTx significantly repressed (about 50%) Oct1-induced MMTV promoter activity, whereas the proteolytically inactive m-LeTx did not repress Oct1-induction of MMTV activity. In addition, the small peptide inhibitor IN-2-LF (L-I1) partially reversed and a combination of inhibitors composed of IN-2-LF, D9R, anti-PA serum and anti-LF serum (L-I2) completely reversed LeTx repression of Oct1-induced MMTV activity. Similar results were found for c-Jun (Fig. 3b) and HNF3 (Fig. 3c). These data indicate that the proteolytic function of LF is required for repression of transcription factor-mediated MMTV activation and that LeTx inhibitors, or mutant LeTx can block this repression.
Fig 3. LeTx inhibitors and the protease deficient mutant of LF reverse LeTx repression of gene activity.
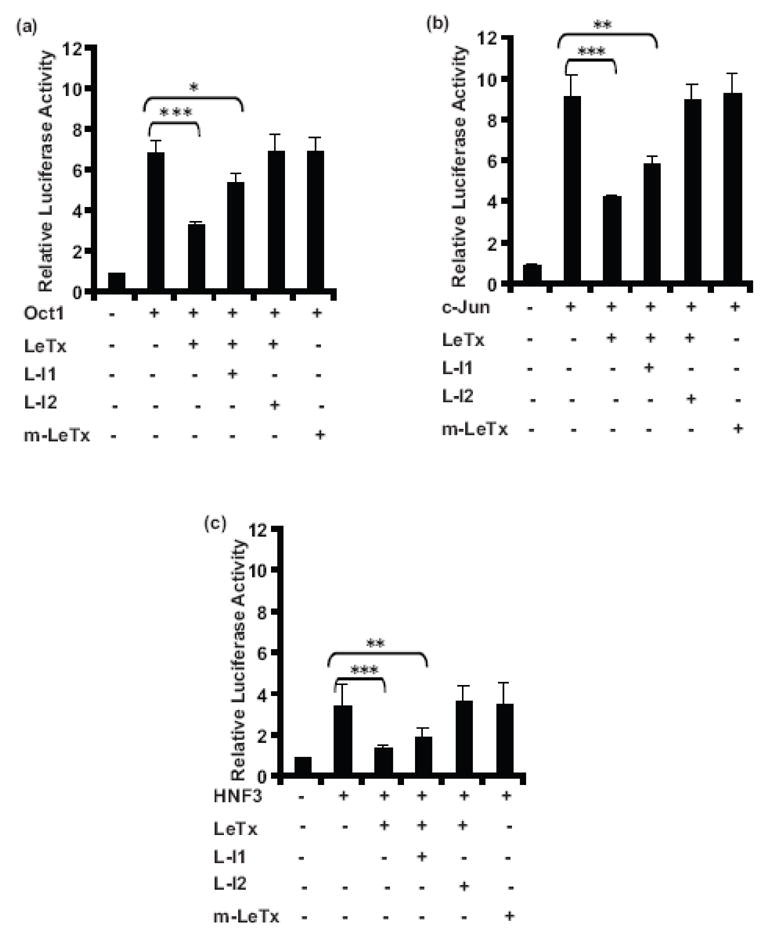
Cos7 cells plated in 24-well plates were transfected 100 ng pGL3-LTR-luc, 15 ng phRL-TK, 40 ng of Oct1 (a), c-Jun (b) or HNF3 (c) expression plasmids using FuGENE6. Cells were treated with wild type LeTx (10 ng/ml LF + 500 ng/ml PA) or m-LeTx (10 ng/ml mutant LF E687C + 500 ng/ml PA) and 10 μM LeTx inhibitor IN-2-LF (L-I1) or a group of LeTx inhibitors (L-I2) (10 μM IN-2-LF + 50 μg/ml anti-PA serum + 50 μg/ml anti-LF serum + 10 μM (D-Arg)9-NH2 peptide) as depicted. After 24 h cells were lysed, and luciferase assayed. Each treatment was performed in triplicates. Means and standard deviations of 3 separate experiments are shown. A one-way ANOVA followed by Tukey’s Multiple Comparison Test was performed (*, p = 0.01–0.05; **, p = 0.001–0.01; ***, p ≤0.001).
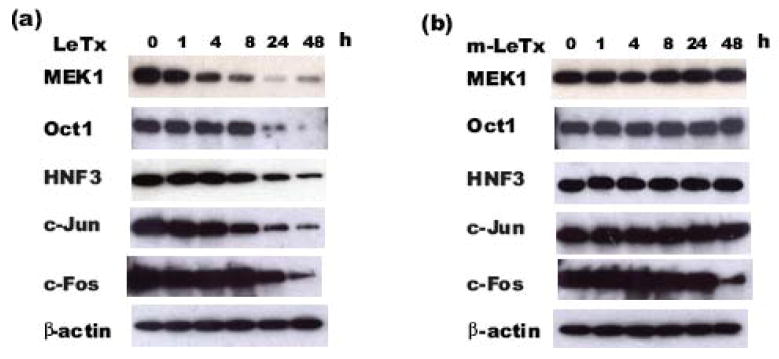
LeTx decreases transcription factor protein levels
Since proteolytic activity is required for LeTx repression of transcription factor induction of the MMTV promoter, the effect of LeTx on the protein levels of these transcription factors was determined. Cos7 cells were transfected and treated following the same protocols as described for Fig. 1. The time course of the protein expression levels of the various transcription factors was measured. We found a significant expression of these transcription factors at 48 hours post-transfection (data not shown). For subsequent experiments, transfections were performed and at 48 hours post-transfection, the media was changed and the cells were then treated with wild type LeTx (10 ng/ml LF + 500 ng/ml PA) or m-LeTx (10 ng/ml mutant LF (E687A) + 500 ng/ml PA) for various lengths of time. As shown in Fig. 4a, LeTx treatment resulted in a decrease in protein levels of the transcription factors Oct1, HNF3, c-Jun and c-Fos in a time-dependent manner. When compared to the cleavage of MEK1, a known LeTx target, the decrease in protein levels of these transcription factors occurred at a relatively late stage after LeTx treatment. Specifically, a small decrease in MEK1 was seen at 1 hour, followed by a dramatic decrease at 4 and 8 hours after LeTx treatment. In contrast, the decrease of these other transcription factors was observed at 8 h (HNF3 and c-Jun) or at 24 h (Oct1, c-Fos) after LeTx treatment. No decrease in MEK1, Oct1, HNF3 or c-Jun protein levels were found in the samples treated with the proteolytic inactive m-LeTx (Fig. 4b). A decrease in c-Fos following 48 h treatment of m-LeTx was observed suggesting that this late time point decrease in c-Fos is independent of the proteolytic activity of LeTx. These data indicate that LeTx decreases protein levels of MEK1, Oct1, HNF3 and c-Jun in a time-dependent manner, which is dependent on the proteolytic activity of LeTx.
Fig 4. LeTx reduces the protein levels of transcription factors.
Cos7 cells plated in 60 mm cell culture plates were transfected with 700 ng pGL3-LTR-luc, 105 ng phRL-TK, 315 ng pSG5 in combination with 280 ng Oct1, HNF3, c-Jun, or c-Fos expression plasmids. 48 h after transfection, Cos-7 cells were incubated in fresh culture media containing wild type LeTx (10 ng/ml LF + 500 ng/ml PA) or m-LeTx (10 ng/ml mutant LF (E687C) + 500 ng/ml PA). At indicated time points after treatments, cells were harvested and total cell lysates were subjected to SDS-PAGE and western blotting using specific antibodies recognizing Oct1, HNF3, c-Jun or c-Fos. The membrane was stripped and re-probed with anti-MEK1 antibody, and then re-probed with anti-β-actin antibody. The representative data from 3 repeats of MEK1, Oct1 and HNF3, and 2 repeats of c-Jun and c-Fos protein levels in LeTx treated samples (a) and m-LeTx treated samples (b) are shown.
The effect of LeTx on endogenous AP-1
The data described are based on over-expression of transcription factors. In order to test the effect of LeTx on endogenous transcription factors, endogenous AP-1 induction of the MMTV promoter and of c-Jun protein was utilized. Cos7 cells were transfected with the MMTV promoter reporter and treated with PMA in the presence or absence of LeTx. The effect of LeTx on PMA-induced endogenous AP-1 activation of MMTV was determined. As shown in Fig. 5a, 10 nM PMA significantly activated the MMTV promoter though endogenous AP-1 and this activity was repressed by LeTx to the basal level. In addition, as shown in Fig. 5b, 24 h treatment of PMA induced an increase in c-Jun protein levels. This increase in PMA-stimulated c-Jun protein levels was largely blocked by treatment with LeTx. Unlike the c-Jun protein, c-Fos protein was undetectable in these cells even after the stimulation of PMA. These data show that LeTx represses PMA-induced endogenous AP-1 activation of MMTV promoter activity and in addition, also inhibits PMA-induced induction of endogenous c-Jun protein levels.
Fig 5. LeTx blocks PMA-induced transcription activity of endogenous AP-1 on the MMTV promoter and PMA-induced endogenous c-Jun protein.
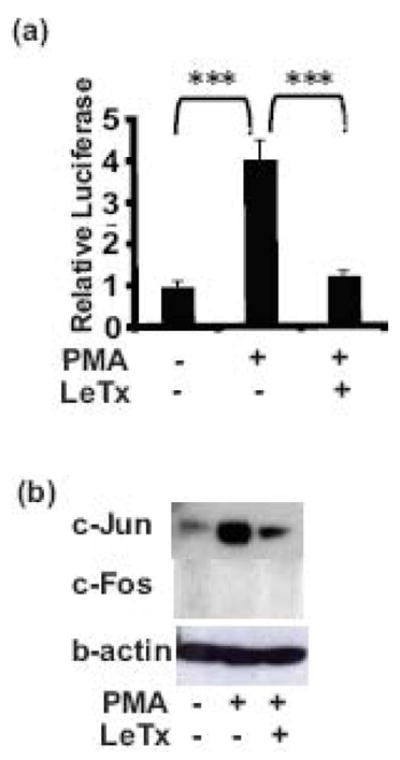
(a). Cos7 cells plated in 24-well plates were transfected with 100 ng pGL3-LTR-luc, 15 ng phRL-TK using FuGENE6. After 24 h incubation in media containing vehicle or 10 nM PMA with/without LeTx (10 ng/ml LF + 500 ng/ml PA), the cells were lysed and luciferase assayed. Each treatment was performed in triplicates. Means and standard deviations of 2 separate experiments are shown. A one-way ANOVA followed by Tukey’s Multiple Comparison Test was performed (*, p = 0.01–0.05; **, p = 0.001–0.01; ***, p ≤0.001). (b). Cos7 cells plated in 60 mm cell culture plates were incubated in starvation conditions (0.5% serum) for 24 h, the cells were treated with vehicle or 10 nM PMA with/without LeTx (10 ng/ml LF + 500 ng/ml PA) in fresh media for 24 h before harvesting for western blotting to detect the c-Jun and c-Fos proteins. The membrane was stripped and re-probed with anti-β-actin antibody. Representative western blot images of c-Jun, c-Fos and β-actin from 3 independent experiments are shown.
Discussion
We have recently shown that LeTx selectively represses nuclear hormone receptors including GR and that LeTx prevents GR-DNA binding in vitro without loss of GR numbers or loss of nuclear translocation, suggesting that other accessory proteins might be the targets for LeTx.16; 17 The studies we report here indicate that LeTx indeed interferes with the action of several transcription factors, including Oct1, HNF3 and cJun. In addition, we found that the protein levels for these transcription factors were decreased at later time points in the presence of fully functional LeTx but not in the presence of a protease inactive mutant, indicating that the reduced function of these transcription factors is dependent on LeTx protease activity.
The LTR of the MMTV promoter, which we have previously shown to be repressed by LeTx.16; 17 contains numerous binding sites for many transcription factors.34–37; 45–47 These transcription factors have been shown to interact with GR and alter GR–induced MMTV promoter activity through influencing the chromatin structure of the promoter. For example, HNF3 has been shown to associate with GR,48 facilitate GR-DNA binding49–52 and alter the chromatin structure of the MMTV LTR.34 Oct1, is able to stimulate glucocorticoid-dependent MMTV promoter activity53–56 and induces chromatin conformational changes of the MMTV promoter.38; 53 c-Jun and its family members show either inhibition or augumentation of GR-mediated transcription of the MMTV-LTR depending on the cell- specific microenvironment.57
In the current study we have found that over-expression of HNF3, Oct1, c-Jun or AP-1 (c-Jun/c-Fos) (Fig. 1) enhanced the basal transcriptional activity of the MMTV promoter. In agreement with previous reports,58; 59 PMA enhanced AP-1-mediated MMTV promoter activity 4 to 8 folds (Fig. 2). LeTx repressed the activation of the MMTV promoter by these transcription factors (Fig. 1) and PMA (Fig. 2). The findings of LeTx repression of MMTV promoter activation through multiple transcriptional factors suggest that the greater extent of repression of a complex promoter such as the MMTV LTR by LeTx, as we have previously described,16; 17 might be due to the synergistic effects of LeTx on multiple transcriptional factors. This is consistent with previous reports of synergy between GR and other transcription factors, as described above.
Since the lethal activity of LeTx is largely dependent on the proteolytic activity of LF,8 we also tested whether the proteolytic function of LF is essential for LeTx repression of transcription factor-mediated activation of the MMTV promoter. The proteolytically inactive mutant LeTx (E687C LF + PA) (m-LeTx)7 failed to repress the transcription factor-mediated MMTV activation (Fig. 3). In addition, we found that a combination of several types of LeTx inhibitors, including a small peptide substrate of LF (IN-2-LF),7; 39–41 a inhibitor of furin (D9R- (D-Arg)9-NH2)40 and anti-serum against PA or LF42–44 completely blocked the repression of MMTV activation by LeTx. These findings suggest that the proteolytic function of LF is essential for its repression of transcription factor-mediated MMTV activation and that a combination of inhibitors functioning at various stages of the LeTx pathway can effectively block LeTx effects.
Much of the research on LeTx has focused on LeTx proteolytic cleavage of MKKs/MEKs.7; 60–62 Recent studies using microarrays and proteomics have shown that LeTx perturbs many genes including transcription factors such as c-Jun, early growth response factor (EGR) 1, 2, 3, Forkhead box c1, and members of the nuclear receptor subfamily 463 and elongation factor 2.14 In the current studies, we show that LeTx not only represses transcription factor-mediated activation of the MMTV promoter (Fig. 3) but also reduced the protein levels of these transcription factors (Fig. 4 and 5). This suggests that the repression could be explained by the reduction of proteins by LeTx. Our findings in the current study and the studies described above have expanded the repertoire of potential LeTx targets.
It is also noteworthy that the repression we report here occurs relatively late after exposure to LeTx: at 24 to 48 hours. This is in contrast to most LeTx studies performed to date, which are based on models observing the early responses of cells or animals within several hours.7; 60–62 In our current study (Fig. 4.), the cleavage of MAPKK is relatively quick (1–4 h). The decrease in Oct1, HNF3 and c-Jun was observed at a later time frame, 8 h or later after LeTx treatment. In agreement with our time course of degradation, it was recently reported that LeTx suppresses c-Jun protein levels at 6 and 12 hr post LeTx treatments in primary endothelial cells.64 The mechanism of the different time course patterns of MEK1 and other transcription factors after LeTx treatment is unclear. However, it is clear that the decrease in the protein levels of these transcription factors in our current study is dependent on LeTx protease activity, since m-LeTx (LF (E687C) + PA) did not reduce the levels of these proteins (Fig. 4b.). It remains to be determined whether the decrease of these transcription factors is a result of direct degradation by LeTx or if it occurs indirectly through unknown mediators. Recently, it was shown that LeTx exposure resulted in the lower mRNA levels of c-Jun and EGRs.63 Reduced IL-8 protein levels were shown to be mediated through mRNA destabilization.65 The late reduction of transcription factor protein levels, which we report here, might also involve indirect effects of LeTx, e.g. lower mRNA levels through lack of de novo mRNA transcription or mRNA destabilization. These findings may shed light on the mechanisms of toxic effects of LeTx at later stages of disease
These transcription factors are also inducible by various stimuli. For instance, c-Jun is activated by JNK,67 TPA,68 PDCD4,69 MAPK,67 and cytokines.70 In this study, as previously reported, we found that PMA induces endogenous AP-1 activation of the MMTV promoter activity (Fig. 5a.), and the induction of the MMTV promoter activity is accompanied by increased protein levels of endogenous c-Jun protein levels (Fig. 5b.). We show here that PMA-induced MMTV activity and PMA-induced endogenous c-Jun protein are repressed by LeTx. These results show that LeTx not only represses over-expressed, but also endogenously expressed transcription factors.
Interestingly, c-Fos protein was not detected in these cells. We also found that transient expression of c-Fos protein alone did not affect the activity of the MMTV promoter (data not shown), suggesting that c-Fos induction of the MMTV promoter may be cell specific. This is consistent with previous reports of the cell-specific expression, function and response of c-Jun and c-Fos to PMA stimulation.57
Importantly, these transcription factors play an essential role in determining the fate of a cell by affecting the expression of target genes involved in proliferation, differentiation and a wide variety of cellular functions. For example, AP-1 is involved in inflammation and apoptosis;71–73 Oct1 has been implicated in the regulation of histones, small nuclear RNAs interleukins and immunoglobulins.74–77 HNF3/fork head transcription factor families play a pivotal role in the regulation of metabolism-related genes.78; 79 In addition, knockout animals have shown the importance of these transcription factors. Knockouts of HNF3α80 or c-Jun81 are incompatible with life. Knockouts of c-Fos81; 82 are viable, but with various deficiencies, e.g. in the stress response,83 bone cell differentiation and bone development and remodeling,81; 84; 85 Taking into consideration such important biological functions of transcription factors, our findings on LeTx repression of these transcription factors could be significant in anthrax lethality.
Materials and methods
Materials
The recombinant B. anthracis toxin proteins LF and PA were kindly provided by S. H. Leppla (NIAID, Bethesda, MD). The pCMV6-XL6 expression plasmids of transcription factor HNF3, Oct1, c-Jun, c-Fos were purchased from Origene (Rockville, MD, USA). Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture
Cos7 cells were grown at 37 °C and 5% CO2 in DMEM containing 10% fetal calf serum, 100 IU/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine.
Construction of full-length MMTV reporter, pGL3-MMTV-luc
The MMTV LTR promoter region containing nucleotides (−1178 to +107) was amplified from pLTR-luc reporter plasmid 30 using primers containing a Nhe I or XhoI site, (−1178) 5′-tcttatGCTAGCcgcctgcagcagaaatggttg-3′ and 5′-tcttatCTCGAGgggtctgcggggggaccctct -3′ (+107). A PCR regime of 98 °C for 5 min followed by 22 cycles of 98 °C for 1 min, 66 °C for 1 min and 72 °C for 1.5 min and a final extension of 72 °C for 7 min was performed using Deep Vent DNA Polymerase (New England Biolabs, Ipswich, MA). The PCR product and receiving vector pGL3-basic vector (Promega, Madison, WI) were digested for 1 h at 37 °C with Nhe I and Xho I, run on a 1% agarose gel. The PCR products were purified using QIAquick gel extraction kit (QIAGEN, Valencia, CA), ligated into lineralized vectors using a Quick Ligation Kit (New England Biolabs) and transfected into DH5a cells (Invitrogen, Carlsbad, CA). pGL3-MMTV-luc was confirmed by restriction enzyme digest and sequencing.
Transient Transfections
Cos7 cells were plated in 24-well plates at a density of 1.5 × 105 cells/well in DMEM containing 10% charcoal-stripped fetal calf serum (cs-FCS), 100 IU/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine 1 day prior to transfection. Cos7 cells were transfected overnight with 100 ng of the reporter construct pGL3-MMTV-luc, 15 ng of the constitutive phRL-TK (Promega), in the absence or presence of various expression plasmids for the transcription factors HNF3 (FOXA3), Oct1 (POU2F1), c-Jun, c-Fos (AP-1), using FuGENE6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. The total amount of DNA per transfection was normailzed to 200 ng using the empty vector pSG5 (Stratagene, La Jolla, CA). The media was replaced with fresh DMEM containing 10% cs-FBS, in the presence or absence of LeTx (10 ng/ml LF + 500 ng/ml PA) and/or specific amounts of LeTx inhibitors and other chemicals as indicated. After 24 h, the cells were lysed, and the firefly and renilla luciferases were assayed using the dual luciferase assay (Promega).
Western blot Analysis
Whole cell extracts were prepared using M-PER (Pierce, Rockford, IL) following the manufacturer’s instructions. The total protein concentrations were determined using a BCA protein assay kit (Pierce). Samples were denatured by heating at 95 °C for 5 min in SDS protein gel loading solution containing 2/5% 2-mercaptoethanol (Sigma-Aldrich, St. Louis). 20 μg of protein was loaded onto a precast 10% acrylamide gel (Biorad Laboratories Inc., Hercules, CA) and run at constant voltage at 200 V for 1 h. Samples were transferred to PVDF by wet blotting at constant voltage of 100 V for 1 h. Membranes were blocked for 1 h at room temperature in 5% fat-free milk and then incubated overnight 4 °C with the primary antibody in 1% fat-free-milk. Dilutions and primary antibodies used are listed in Table 1.
Table 1.
Primary antibody
| Primary Ab | Catalog No. | Vender | Dilution | Size |
|---|---|---|---|---|
| HNF3 | PA1-25391 | Affinity BioReagents (Golden, CO) | 1:1300 | 43 kDa |
| Oct1 | Ab51363 | Abcam Inc (Cambridge, MA) | 1:125 | 80 kDa |
| c-Jun | 9165 | Cell Signaling Technology (Danvers, MA) | 1:1500 | 41 kDa |
| c-Fos | 2250 | Cell Signaling Technology (Danvers, MA) | 1:2000 | 57/62 kDa |
| MEK-1 | 07-641 | Millipore (Billerica, MA) | 1:1500 | 45 kDa |
| β-actin | sc-47778 | Santa Cruz Biotechnology Inc (Santa Cruz, CA) | 1:2500 | 42 kDa |
Membranes were then washed 3 × 10 min with TBST (TBS containing 0.1% Tween-20) and incubated with an HRP-conjugated specific secondary antibody (Pierce) plus HRP-conjugated anti-biotin antibody at room temperature for 1 h. Membranes were again washed 3 × 10 min with TTBS and bands detected using the Supersignal West Dura Extended Duration Substrate (Pierce).
Statistical analysis
Statistical analysis of the data was performed using one-way or two-way ANOVA and an appropriate post-hoc test.
Acknowledgments
This work was supported by the NIMH/NIH Intramural Research Program and an NIAID/NIH Intramural Research Program Biodefense Research Grant.
We thank Stephen Leppla for providing the anthrax lethal toxin and Gordon Hager for plasmid. We would also like to thank Mahtab Moayeri for helpful discussions and Marni Silverman for assisting with the preparation of the manuscript.
Abbreviations
- LeTx
anthrax lethal toxin
- m-LeTx
mutant anthrax lethal toxin
- LF
lethal factor
- EF
edema factor
- PA
protective antigen
- MAPKK
mitogen activated protein kinase kinase
- GR
glucocorticoid receptor
- HPA
hypothalamic-pituitary–adrenal
- MMTV
mouse mammary tumor virus
- GRE
glucocorticoid response element
- LTR
long terminal repeat
- HNF3
hepatocyte nuclear factor 3
- Oct1
octamer-binding protein 1
- AP-1
activator protein 1
- PMA
Phorbol 12-myristate 13-acetate
- cs-FCS
charcoal-stripped fetal calf serum
- EGR
early growth response factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhatnagar R, Batra S. Anthrax toxin. Crit Rev Microbiol. 2001;27:167–200. doi: 10.1080/20014091096738. [DOI] [PubMed] [Google Scholar]
- 2.Collier RJ, Young JA. Anthrax toxin. Annu Rev Cell Dev Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 3.Leppla SH. Anthrax Toxin. In: Aktories KaJI., editor. Bacterial Protein Toxins. Springer; Berlin: 2000. pp. 445–472. [Google Scholar]
- 4.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem. 2007;76:243–65. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 5.Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55:647–71. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 6.Mourez M, Lacy DB, Cunningham K, Legmann R, Sellman BR, Mogridge J, Collier RJ. 2001: a year of major advances in anthrax toxin research. Trends Microbiol. 2002;10:287–93. doi: 10.1016/s0966-842x(02)02369-7. [DOI] [PubMed] [Google Scholar]
- 7.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–7. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 8.Klimpel KR, Arora N, Leppla SH. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol Microbiol. 1994;13:1093–100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. Anthrax lethal factor cleaves MKK3 in macrophages and inhibits the LPS/IFNγ-induced release of NO and TNFα. FEBS letters. 1999;462:199–204. doi: 10.1016/s0014-5793(99)01502-1. [DOI] [PubMed] [Google Scholar]
- 10.Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. Lethal factor of Bacillus anthracis cleaves the N-terminus of MAPKKs: analysis of the intracellular consequences in macrophages. Int J Med Microbiol. 2000;290:421–7. doi: 10.1016/S1438-4221(00)80056-9. [DOI] [PubMed] [Google Scholar]
- 11.Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Susceptibility of mitogen-activated protein kinase kinase family members to proteolysis by anthrax lethal factor. Biochem J. 2000;352(Pt 3):739–45. [PMC free article] [PubMed] [Google Scholar]
- 12.Welkos SL, Keener TJ, Gibbs PH. Differences in Susceptibility of Inbred Mice to Bacillus anthracis. Infect Immun. 1986;51:795–800. doi: 10.1128/iai.51.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comer JE, Galindo CL, Chopra AK, Peterson JW. GeneChip analyses of global transcriptional responses of murine macrophages to the lethal toxin of Bacillus anthracis. Infect Immun. 2005;73:1879–85. doi: 10.1128/IAI.73.3.1879-1885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapra R, Gaucher SP, Lachmann JS, Buffleben GM, Chirica GS, Comer JE, Peterson JW, Chopra AK, Singh AK. Proteomic analyses of murine macrophages treated with Bacillus anthracis lethal toxin. Microb Pathog. 2006;41:157–67. doi: 10.1016/j.micpath.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Moayeri M, Leppla SH. The roles of anthrax toxin in pathogenesis. Curr Opin Microbiol. 2004;7:19–24. doi: 10.1016/j.mib.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Webster JI, Sternberg EM. Anthrax lethal toxin represses glucocorticoid receptor (GR) transactivation by inhibiting GR-DNA binding in vivo. Mol Cell Endocrinol. 2005;241:21–31. doi: 10.1016/j.mce.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Webster JI, Tonelli LH, Moayeri M, Simons SS, Jr, Leppla SH, Sternberg EM. Anthrax lethal factor represses glucocorticoid and progesterone receptor activity. Proc Natl Acad Sci U S A. 2003;100:5706–11. doi: 10.1073/pnas.1036973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards CKI, Yunger LM, Lorence RM, Dantzer R, Kelley KW. The pituitary gland is required for protection against lethal effects of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:2274–2277. doi: 10.1073/pnas.88.6.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez SA, Fernandez GC, Vanzulli S, Dran G, Rubel C, Berki T, Isturiz MA, Palermo MS. Endogenous glucocorticoids attenuate Shiga toxin-2-induced toxicity in a mouse model of haemolytic uraemic syndrome. Clin Exp Immunol. 2003;131:217–224. doi: 10.1046/j.1365-2249.2003.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacPhee IAM, Antoni FA, Mason DW, et al. Spontaneous recovery of rats from experimental allergic encephalomyelitis is dependent on regulation of the immune system by endogenous adrenal corticosteroids. J Exp Med. 1989;169:431–445. doi: 10.1084/jem.169.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moayeri M, Webster JI, Wiggins JF, Leppla SH, Sternberg EM. Endocrine perturbation increases susceptibility of mice to anthrax lethal toxin. Infect Immun. 2005;73:4238–44. doi: 10.1128/IAI.73.7.4238-4244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzek MC, Pearce BD, Miller AH, Biron CA, et al. Endogenous Glucocorticoids Protect Against Cytokine-Mediated Lethality During Viral Infection. J Immunol. 1999;162:3527–3533. [PubMed] [Google Scholar]
- 23.Sternberg EM, Hill JM, Chrousos GP, Kamilaris T, Listwak SJ, Gold PW, Wilder RL. Inflammatory mediator-induced hypothalamic-pituitary-adrenal axis activation is defective in streptococcal cell wall arthritis- susceptible Lewis rats. Proc Natl Acad Sci U S A. 1989;86:2374–8. doi: 10.1073/pnas.86.7.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster JI, Sternberg EM. Role of the hypothalamic-pituitary-adrenal axis, glucocorticoids and glucocorticoid receptors in toxic sequelae of exposure to bacterial and viral products. J Endocrinol. 2004;181:207–21. doi: 10.1677/joe.0.1810207. [DOI] [PubMed] [Google Scholar]
- 25.Zuckerman SH, Qureshi N. In vivo inhibition of lipopolysaccharide-induced lethality and tumor necrosis factor synthesis by Rhodobacter sphaeroides diphosphoryl lipid A is dependent on corticosterone induction. Infect Immun. 1992;60:2581–7. doi: 10.1128/iai.60.7.2581-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinyamu HK, Archer TK. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim Biophys Acta. 2004;1677:30–45. doi: 10.1016/j.bbaexp.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 27.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 28.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 29.Geisse S, Scheidereit C, Westphal HM, Hynes NE, Groner B, Beato M. Glucocorticoid receptors recognize DNA sequences in and around murine mammary tumour virus DNA. Embo J. 1982;1:1613–9. doi: 10.1002/j.1460-2075.1982.tb01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebvre P, Berard DS, Cordingley MG, Hager GL. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol Cell Biol. 1991;11:2529–37. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce J, Fee BE, Toohey MG, Peterson DO. A mouse mammary tumor virus promoter element near the transcription initiation site. J Virol. 1993;67:415–24. doi: 10.1128/jvi.67.1.415-424.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–6. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 33.Bresnick EH, John S, Berard DS, LeFebvre P, Hager GL. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci U S A. 1990;87:3977–81. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmqvist PH, Belikov S, Zaret KS, Wrange O. FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp Cell Res. 2005;304:593–603. doi: 10.1016/j.yexcr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Beato M. Transcriptional control by nuclear receptors. Faseb J. 1991;5:2044–51. doi: 10.1096/fasebj.5.7.2010057. [DOI] [PubMed] [Google Scholar]
- 36.Truss M, Bartsch J, Mows C, Chavez S, Beato M. Chromatin structure of the MMTV promoter and its changes during hormonal induction. Cell Mol Neurobiol. 1996;16:85–101. doi: 10.1007/BF02088169. [DOI] [PubMed] [Google Scholar]
- 37.Herrlich P. Cross-talk between glucocorticoid receptor and AP-1. Oncogene. 2001;20:2465–75. doi: 10.1038/sj.onc.1204388. [DOI] [PubMed] [Google Scholar]
- 38.Belikov S, Astrand C, Holmqvist PH, Wrange O. Chromatin-mediated restriction of nuclear factor 1/CTF binding in a repressed and hormone-activated promoter in vivo. Mol Cell Biol. 2004;24:3036–47. doi: 10.1128/MCB.24.7.3036-3047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Choi MK, Koo BS, Yoon MY. Development of high-throughput assay of lethal factor using native substrate. Anal Biochem. 2005;341:33–9. doi: 10.1016/j.ab.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Peinado JR, Kacprzak MM, Leppla SH, Lindberg I. Cross-inhibition between furin and lethal factor inhibitors. Biochem Biophys Res Commun. 2004;321:601–5. doi: 10.1016/j.bbrc.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Turk BE, Wong TY, Schwarzenbacher R, Jarrell ET, Leppla SH, Collier RJ, Liddington RC, Cantley LC. The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Nat Struct Mol Biol. 2004;11:60–6. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 42.Kobiler D, Gozes Y, Rosenberg H, Marcus D, Reuveny S, Altboum Z. Efficiency of protection of guinea pigs against infection with Bacillus anthracis spores by passive immunization. Infect Immun. 2002;70:544–60. doi: 10.1128/IAI.70.2.544-550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staats HF, Alam SM, Scearce RM, Kirwan SM, Zhang JX, Gwinn WM, Haynes BF. In vitro and in vivo characterization of anthrax anti-protective antigen and anti-lethal factor monoclonal antibodies after passive transfer in a mouse lethal toxin challenge model to define correlates of immunity. Infect Immun. 2007;75:5443–52. doi: 10.1128/IAI.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbull PC, Broster MG, Carman JA, Manchee RJ, Melling J. Development of antibodies to protective antigen and lethal factor components of anthrax toxin in humans and guinea pigs and their relevance to protective immunity. Infect Immun. 1986;52:356–63. doi: 10.1128/iai.52.2.356-363.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funder JW. Mineralocorticoids, glucocorticoids, receptors and response elements. Science. 1993;259:1132–3. doi: 10.1126/science.8382375. [DOI] [PubMed] [Google Scholar]
- 46.Hsu TC, Melchiorre LP, Jr, Maksymowych AB, Kmiec E, Litwack G. Assembly of glucocorticoid receptor and c-JUN homodimer on the promoter of mouse mammary tumor virus-long terminal repeat is influenced by order of addition. Biochem Biophys Res Commun. 1993;197:1260–6. doi: 10.1006/bbrc.1993.2613. [DOI] [PubMed] [Google Scholar]
- 47.Siddique HR, Sarkar NH. The interaction of a c-Jun/Fos related protein factor with the U3 sequences of the mouse mammary tumor virus LTR. Biochem Biophys Res Commun. 1990;172:348–56. doi: 10.1016/s0006-291x(05)80216-5. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T, Scott DK, Wang JC, Granner DK. Structural requirements of the glucocorticoid and retinoic acid response units in the phosphoenolpyruvate carboxykinase gene promoter. Mol Endocrinol. 1998;12:1487–98. doi: 10.1210/mend.12.10.0187. [DOI] [PubMed] [Google Scholar]
- 49.Espinas ML, Roux J, Pictet R, Grange T. Glucocorticoids and protein kinase A coordinately modulate transcription factor recruitment at a glucocorticoid-responsive unit. Mol Cell Biol. 1995;15:5346–54. doi: 10.1128/mcb.15.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stafford JM, Wilkinson JC, Beechem JM, Granner DK. Accessory factors facilitate the binding of glucocorticoid receptor to the phosphoenolpyruvate carboxykinase gene promoter. J Biol Chem. 2001;276:39885–91. doi: 10.1074/jbc.M105370200. [DOI] [PubMed] [Google Scholar]
- 51.Wang JC, Stromstedt PE, O’Brien RM, Granner DK. Hepatic nuclear factor 3 is an accessory factor required for the stimulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Mol Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 52.Wang JC, Stromstedt PE, Sugiyama T, Granner DK. The phosphoenolpyruvate carboxykinase gene glucocorticoid response unit: identification of the functional domains of accessory factors HNF3 beta (hepatic nuclear factor-3 beta) and HNF4 and the necessity of proper alignment of their cognate binding sites. Mol Endocrinol. 1999;13:604–18. doi: 10.1210/mend.13.4.0269. [DOI] [PubMed] [Google Scholar]
- 53.Belikov S, Holmqvist PH, Astrand C, Wrange O. Nuclear factor 1 and octamer transcription factor 1 binding preset the chromatin structure of the mouse mammary tumor virus promoter for hormone induction. J Biol Chem. 2004;279:49857–67. doi: 10.1074/jbc.M409713200. [DOI] [PubMed] [Google Scholar]
- 54.Bruggemeier U, Kalff M, Franke S, Scheidereit C, Beato M. Ubiquitous transcription factor OTF-1 mediates induction of the MMTV promoter through synergistic interaction with hormone receptors. Cell. 1991;64:565–72. doi: 10.1016/0092-8674(91)90240-y. [DOI] [PubMed] [Google Scholar]
- 55.Prefontaine GG, Lemieux ME, Giffin W, Schild-Poulter C, Pope L, LaCasse E, Walker P, Hache RJ. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol Cell Biol. 1998;18:3416–30. doi: 10.1128/mcb.18.6.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toohey MG, Lee JW, Huang M, Peterson DO. Functional elements of the steroid hormone-responsive promoter of mouse mammary tumor virus. J Virol. 1990;64:4477–88. doi: 10.1128/jvi.64.9.4477-4488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maroder M, Farina AR, Vacca A, Felli MP, Meco D, Screpanti I, Frati L, Gulino A. Cell-specific bifunctional role of Jun oncogene family members on glucocorticoid receptor-dependent transcription. Mol Endocrinol. 1993;7:570–84. doi: 10.1210/mend.7.4.8388998. [DOI] [PubMed] [Google Scholar]
- 58.Cho HJ, Kang JH, Kwak JY, Lee TS, Lee IS, Park NG, Nakajima H, Magae J, Chang YC. Ascofuranone suppresses PMA-mediated matrix metalloproteinase-9 gene activation through the Ras/Raf/MEK/ERK- and Ap1-dependent mechanisms. Carcinogenesis. 2007;28:1104–10. doi: 10.1093/carcin/bgl217. [DOI] [PubMed] [Google Scholar]
- 59.Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–31. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 60.Bromberg-White JL, Duesbery NS. Biological and biochemical characterization of anthrax lethal factor, a proteolytic inhibitor of MEK signaling pathways. Methods Enzymol. 2008;438:355–65. doi: 10.1016/S0076-6879(07)38025-7. [DOI] [PubMed] [Google Scholar]
- 61.Chopra AP, Boone SA, Liang X, Duesbery NS. Anthrax lethal factor proteolysis and inactivation of MAPK kinase. J Biol Chem. 2003;278:9402–6. doi: 10.1074/jbc.M211262200. [DOI] [PubMed] [Google Scholar]
- 62.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Anthrax lethal factor cleaves the N-terminus of MAPKKS and induces tyrosine/threonine phosphorylation of MAPKS in cultured macrophages. J Appl Microbiol. 1999;87:288. doi: 10.1046/j.1365-2672.1999.00893.x. [DOI] [PubMed] [Google Scholar]
- 63.Barson HV, Mollenkopf H, Kaufmann SH, Rijpkema S. Anthrax lethal toxin suppresses chemokine production in human neutrophil NB-4 cells. Biochem Biophys Res Commun. 2008;374:288–93. doi: 10.1016/j.bbrc.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 64.Warfel JM, D’Agnillo F. Anthrax lethal toxin enhances TNF-induced endothelial VCAM-1 expression via an IFN regulatory factor-1-dependent mechanism. J Immunol. 2008;180:7516–24. doi: 10.4049/jimmunol.180.11.7516. [DOI] [PubMed] [Google Scholar]
- 65.Batty S, Chow EM, Kassam A, Der SD, Mogridge J. Inhibition of mitogen-activated protein kinase signalling by Bacillus anthracis lethal toxin causes destabilization of interleukin-8 mRNA. Cell Microbiol. 2006;8:130–8. doi: 10.1111/j.1462-5822.2005.00606.x. [DOI] [PubMed] [Google Scholar]
- 66.Jernigan JA, Stephens DS, Ashford DA, Omenaca C, Topiel MS, Galbraith M, Tapper M, Fisk TL, Zaki S, Popovic T, Meyer RF, Quinn CP, Harper SA, Fridkin SK, Sejvar JJ, Shepard CW, McConnell M, Guarner J, Shieh WJ, Malecki JM, Gerberding JL, Hughes JM, Perkins BA. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis. 2001;7:933–44. doi: 10.3201/eid0706.010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan MA, Bouzari S, Ma C, Rosenberger CM, Bergstrom KS, Gibson DL, Steiner TS, Vallance BA. Flagellin-dependent and -independent inflammatory responses following infection by enteropathogenic Escherichia coli and Citrobacter rodentium. Infect Immun. 2008;76:1410–22. doi: 10.1128/IAI.01141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simantov R. Neurotransporters: regulation, involvement in neurotoxicity, and the usefulness of antisense nucleic acids. Biochem Pharmacol. 1995;50:435–42. doi: 10.1016/0006-2952(95)00068-b. [DOI] [PubMed] [Google Scholar]
- 69.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–76. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 70.Yokoo T, Kitamura M. Antioxidant PDTC induces stromelysin expression in mesangial cells via a tyrosine kinase-AP-1 pathway. Am J Physiol. 1996;270:F806–11. doi: 10.1152/ajprenal.1996.270.5.F806. [DOI] [PubMed] [Google Scholar]
- 71.Nomura S, Takano-Yamamoto T. Molecular events caused by mechanical stress in bone. Matrix Biol. 2000;19:91–6. doi: 10.1016/s0945-053x(00)00050-0. [DOI] [PubMed] [Google Scholar]
- 72.Reme CE, Grimm C, Hafezi F, Iseli HP, Wenzel A. Why study rod cell death in retinal degenerations and how? Doc Ophthalmol. 2003;106:25–9. doi: 10.1023/a:1022423724376. [DOI] [PubMed] [Google Scholar]
- 73.Sevilla L, Zaldumbide A, Pognonec P, Boulukos KE. Transcriptional regulation of the bcl-x gene encoding the anti-apoptotic Bcl-xL protein by Ets, Rel/NFkappaB, STAT and AP1 transcription factor families. Histol Histopathol. 2001;16:595–601. doi: 10.14670/HH-16.595. [DOI] [PubMed] [Google Scholar]
- 74.Gruart-Gouilleux V, Engels P, Sullivan M. Characterization of the human interleukin-5 gene promoter: involvement of octamer binding sites in the gene promoter activity. Eur J Immunol. 1995;25:1431–5. doi: 10.1002/eji.1830250544. [DOI] [PubMed] [Google Scholar]
- 75.La Bella F, Heintz N. Histone gene transcription factor binding in extracts of normal human cells. Mol Cell Biol. 1991;11:5825–31. doi: 10.1128/mcb.11.12.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matthias P. Lymphoid-specific transcription mediated by the conserved octamer site: who is doing what? Semin Immunol. 1998;10:155–63. doi: 10.1006/smim.1998.0117. [DOI] [PubMed] [Google Scholar]
- 77.Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–61. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaestner KH. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol Metab. 2000;11:281–5. doi: 10.1016/s1043-2760(00)00271-x. [DOI] [PubMed] [Google Scholar]
- 79.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 80.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–6. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 82.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992;71:577–86. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 83.Kubovcakova L, Kvetnansky R, Krizanova O. [Use of knock-out mice in studies of stress reactions] Cesk Fysiol. 2003;52:118–28. [PubMed] [Google Scholar]
- 84.Boyce BF, Hughes DE, Wright KR, Xing L, Dai A. Recent advances in bone biology provide insight into the pathogenesis of bone diseases. Lab Invest. 1999;79:83–94. [PubMed] [Google Scholar]
- 85.Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61(Suppl 2):ii40–2. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]