Abstract
The initiation of DNA synthesis is thought to occur at sites bound by a heteromeric origin recognition complex (ORC). Previously, we have shown that in Drosophila, the level of the large subunit, ORC1, is modulated during cell cycle progression and that changes in ORC1 concentration alter origin utilization during development. Here, we investigate the mechanisms underlying cell cycle-dependent degradation of ORC1. We show that signals in the non-conserved N-terminal domain of ORC1 mediate its degradation upon exit from mitosis and in G1 phase by the anaphase-promoting complex (APC) in vivo. Degradation appears to be the result of direct action of the APC, as the N-terminal domain is ubiquitylated by purified APC in vitro. This regulated proteolysis is potent, sufficient to generate a normal temporal distribution of protein even when transcription of ORC1 is driven by strong constitutive promoters. These observations suggest that in Drosophila, ORC1 regulates origin utilization much as does Cdc6 in budding yeast.
Keywords: APC/cell cycle/ORC1/proteolysis/replication
Introduction
Control of cell cycle progression underlies the orderly proliferation of cells essential for normal development and homeostasis in adult animals. The initiation of DNA replication commits cells in organisms as diverse as yeast and mammals to progression through a complete cycle. In addition, replication is misregulated in most tumor cells, in part accounting for their phenotype (Sherr, 1996). Therefore, there is a great deal of interest in understanding the molecular mechanisms of replication initiation.
The initiation of DNA synthesis is perhaps best understood in the budding yeast where discrete well-defined sequences that act as origins in vivo were defined some time ago (reviewed by Newlon and Theis, 1993; Bell, 1995). The foundation for mechanistic studies was the identification of a heteromeric hexamer, the origin recognition complex (ORC), that binds to yeast origins (Bell and Stillman, 1992). The ORC marks origins, serving as a platform for subsequent loading of additional factors including Cdc6 (Stillman, 1996; Bogan et al., 2000; Diffley, 2001; Bell, 2002; Bell and Dutta, 2002). In budding yeast, ORCs are monotonous: as cells progress through the cycle, they remain constitutively bound to origins and their levels do not fluctuate even when yeast enter a prolonged quiescent period that is perhaps analogous to the G0 phase of mammalian cells (Aparicio et al., 1997; Liang and Stillman, 1997; Tanaka et al., 1997). Origin utilization is governed by recruitment of Cdc6, the abundance of which is tightly regulated during the cell cycle (for a review see Bell and Dutta, 2002).
ORC proteins are well conserved throughout metazoans, suggesting they might constitutively mark replication origins throughout the cell cycle in other organisms as well. This idea is supported by some studies of mammalian cells (Ritzi et al., 1998; Saha et al., 1998; Tatsumi et al., 2000) and of early Xenopus embryos. The abundance of ORC proteins in these cell types is also constant throughout the cell cycle, although the Xenopus ORCs are released from chromatin during M phase, unlike their budding yeast counterparts (Hua and Newport, 1998; Findeisen et al., 1999; Rowles et al., 1999). However, other work on human cells as well as work on hamster cells and Drosophila has revealed significantly different behavior for the largest subunit, ORC1.
In cycling CHO cells, ORC1, but not other ORC subunits, is released from chromatin at the end of S phase (Natale et al., 2000; Li and DePamphilis, 2002). The behavior of ORC1 in cycling HeLa and Raji human cells is even more divergent from the budding yeast paradigm (Fujita et al., 2002; Mendez et al., 2002). ORC1 levels rise in G1 and fall during S phase. Accumulation in G1 is due, at least in part, to modulated transcription of the orc1 gene by E2F (Ohtani et al., 1996), a conserved transcription factor that plays a central role in orchestrating the G1–S transition. Following S phase entry, two events lead to the disappearance of ORC1. First, E2F activity falls so that degraded ORC1 cannot be replenished. Secondly, the F-box protein Skp2 mediates degradation of ORC1 either while still bound to chromatin or very quickly upon its release, as unbound protein is undetectable (Mendez et al., 2002). The consequences of failing to modulate either the level of ORC1 or its association with chromatin are unclear, although it has been speculated that ORC1 may be a limiting factor for pre-replication complex formation in early G1 (Cimbora and Groudine, 2001; Li and DePamphilis, 2002; Mendez et al., 2002).
ORC1 levels are also modulated in Drosophila, where some of the consequences of misaccumulation are known (Asano and Wharton, 1999). As in human cells, the level of ORC1, but not ORC2 (Pak et al., 1997), is modulated in proliferating cells of the Drosophila embryo or imaginal disc. ORC1 accumulates at the end of G1 as a result of transient E2F-dependent transcription, just as in human cells. Although the precise timing of ORC1 degradation was not determined, the protein disappears sometime after S phase entry but before G1 of the subsequent cycle. Most importantly, misexpression of ORC1 causes an ensemble of phenotypes that include aberrant S phase entry, female sterility and lethality. Taken together, these suggest that at least some of the cells in embryonic, larval, imaginal and adult tissues are extremely sensitive to the level of ORC1 (Asano and Wharton, 1999). Despite its importance, the pathways responsible for cell cycle-coupled degradation of ORC1 have not been described in Drosophila.
In this report, we investigate the timing and mechanism of ORC1 degradation. In eye imaginal disc cells, ORC1 persists from the end of G1 until M phase, disappearing only upon completion of mitosis and entry into the subsequent G1 phase. This finding suggested that ORC1 is degraded by the anaphase-promoting complex (APC), a key component of the cell cycle clock responsible for the cyclical degradation of mitotic cyclins and securin (Morgan, 1999; Zachariae and Nasmyth, 1999; Harper et al., 2002). In support of this idea, we find that ORC1 degradation in vivo is enhanced by overexpression of fizzy-related (fzr), an activating subunit of the APC. This action of Fzr apparently is direct, since we observe Fzr-dependent ubiquitylation of ORC1 by purified APC.
Results
ORC1 degradation during M phase
Previously we have shown that ORC1 accumulates transiently during a synchronous cycle that takes place in third larval instar eye imaginal disc cells (Figure 1A) (Asano and Wharton, 1999). The onset of accumulation appears to result from transient activation of E2F-dependent transcription in the interval from late G1 to early S phase. Due to the methods of detection in these experiments, it was not possible to determine precisely when during the cycle ORC1 disappeared. In this report, we have used confocal microscopy to examine the timing of ORC1 disappearance with respect to two cell cycle markers: cyclin B (CycB), which accumulates from S phase until its destruction by the APC during mitosis (Lehner and O’Farrell, 1990; Whitfield et al., 1990; Sigrist et al., 1995; Huang and Raff, 1999), and the phospho-histone 3 (PH3) epitope, which marks M phase.
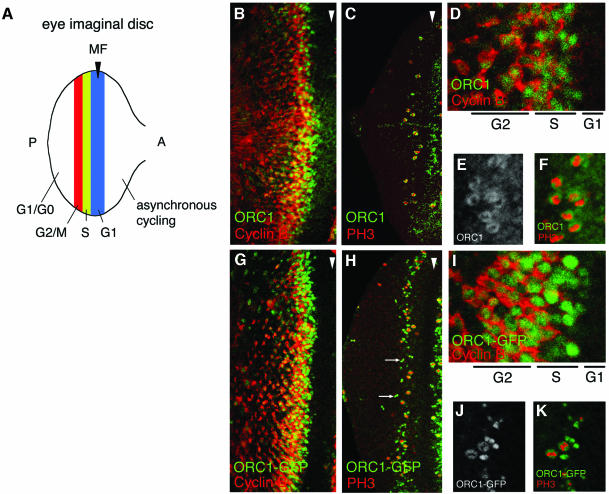
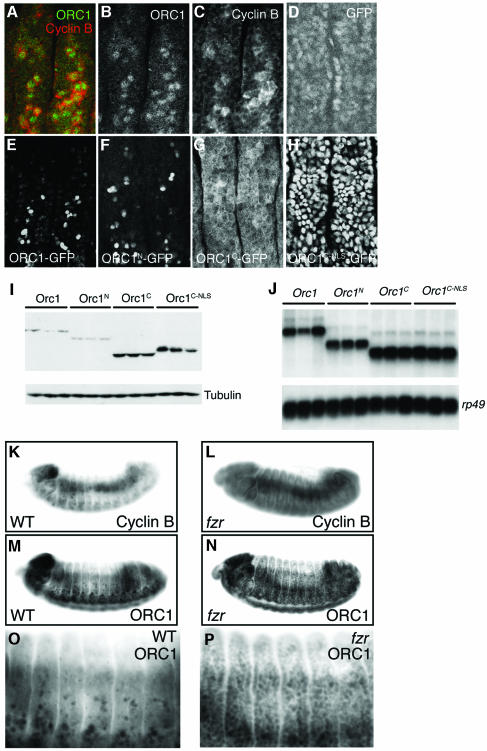
Fig. 1. Persistence of ORC1 into M phase. (A) Schematic of the synchronous cell cycle transition in the eye imaginal disc. As the morphogenetic furrow (MF, hereinafter marked with an arrowhead) sweeps from posterior (P) to anterior (A), most cells undergo a synchronous transition and then enter a prolonged G1/G0 phase. (Note that some cells behind the MF are in a prolonged G2 arrest, as shown in Figure 3.) Ahead of the furrow and in the attached antennal disc, cells cycle asynchronously. (B–K) Confocal images of eye imaginal discs near the MF (arrowhead). Endogenous ORC1 (B–F) and ORC1–GFP expressed under ORC1 transcriptional control (G–K), and CycB or PH3, as indicated. Interphase nuclei are visible in optical sections through the middle of the disc (B, D, G and I), whereas mitotic nuclei are visible in apical optical sections (C, E, F, H, J and K). Arrows in (H) are examples of late telophase nuclei (see also Figure 3K–M).
As shown in Figure 1B and D, ORC1 is first detectable in the nuclei of cells that have not yet accumulated appreciable cytoplasmic CycB, consistent with our previous determination that ORC1 accumulates just before S phase entry. Examination of serial optical sections (not shown) reveals that both proteins persist in cells as they transit G2. As the cells approach M phase, their nuclei rise apically and accumulate PH3 upon M phase entry (Figure 1C). Remarkably, at this stage of the cell cycle, ORC1 is distributed in a ‘donut’ that surrounds the PH3 (Figure 1E and F), presumably because it has dissociated from the chromatin. Subsequently, both PH3 and ORC1 disappear as the cells complete mitosis and enter G1 of the following cycle.
Our ability to monitor endogenous ORC1 in the experiments described above is limited by the level of antigen and the sensitivity of the antibody. In part to overcome these limitations and in part to monitor the regulation of various ORC1 derivatives, we generated a transgene that encodes a fusion of ORC1 to an epitope-tagged green fluorescent protein (GFP) derivative and expressed the resulting fusion protein under control of the native orc1 promoter. Transgenic flies were generated by standard methods, and we compared the distributions of endogenous ORC1 and transgenic ORC1–GFP.
In general, the patterns of endogenous ORC1 and transgenic ORC1–GFP are indistinguishable in ovaries, embryos and the wing imaginal disc (data not shown, see Asano and Wharton, 1999). In addition, detailed examination of the synchronous cell cycle transition in the eye disc described above reveals essentially identical behavior of ORC1 and ORC1–GFP: accumulation of ORC1–GFP prior to CycB in late G1 or early S phase, persistence of both ORC1–GFP and CycB throughout G2, removal of ORC1–GFP from chromatin during M phase upon accumulation of PH3, and disappearance of all three antigens (ORC1–GFP, CycB and PH3) upon entry into the subsequent G1 (Figure 1G–K). Thus, addition of the GFP-myc tag does not appear to perturb the cell cycle-modulated accumulation of ORC1 significantly.
We next wished to determine whether ORC1 accumulation is regulated in a similar fashion in the asynchronously cycling cells that make up most of the imaginal discs. This seemed likely, given the appearance of ORC1 and ORC1–GFP ‘donuts’ in the asynchronous mitotic cells immediately anterior to the morphogenetic furrow in Figure 1. However, to survey the entire population of disc cells, we turned to the fluorescence-activated cell sorting (FACS) protocol devised by Edgar and colleagues. In brief, imaginal discs from transgenic animals expressing ORC1–GFP under orc1 promoter control were collected, dissociated into individual cells, and sorted according to both DNA content and green fluorescence. We examined separately the wing disc, which has been characterized in great detail by this method (Neufeld et al., 1998; Datar et al., 2000), and the eye antennal disc complex.
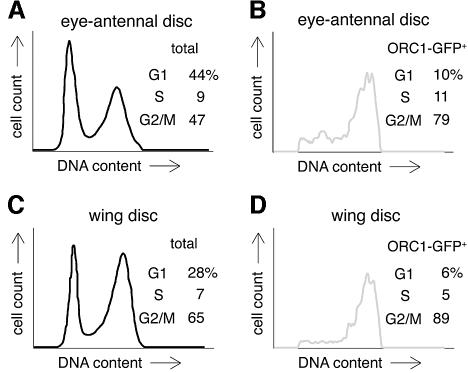
The cell cycle distribution of GFP-positive cells agrees with the analysis described above and in Figure 1: ORC1–GFP is found predominantly in cells with S and G2 DNA content (Figure 2), consistent with the idea that it accumulates near the G1/S boundary, persists through G2, and disappears during M phase. During this stage of development, the eye and wing discs are governed by very different developmental programs; thus, the essentially identical regulation of ORC1 in these two tissues (Figure 2) most probably reflects regulation by intrinsic cell cycle machinery rather than by developmental cues.
Fig. 2. Absence of ORC1 in G1 phase. FACS analysis of dissociated imaginal disc cells from transgenic animals expressing ORC1–GFP under ORC1 transcriptional control. The proportion of G1 cells in eye antennal discs is higher than in wing discs, due to the contribution of terminally differentiating cells behind the morphogenetic furrow.
In summary, the ORC1 in imaginal discs persists from late G1 into M phase when most of the protein is abruptly degraded before entry into the subsequent G1.
Proteolysis epistatic to transcriptional control in modulating ORC1 levels: degradation during G1
The results described above are consistent with the idea that ORC1 accumulates at the G1/S boundary immediately after a pulse of E2F-dependent transcription and persists until its catastrophic destruction at the M/G1 boundary. In this scenario, cell cycle-modulated transcription and proteolysis both contribute to setting the level of ORC1. We wished to test this idea directly by uncoupling the expression of orc1 mRNA from its normal transcriptional signals. To this end, we drove constitutive expression of ORC1–GFP using the GMR promoter, which is turned on in all cells posterior to the morphogenetic furrow in the eye disc (Moses and Rubin, 1991), and then visualized the distribution of green fluorescence both in situ and in FACS experiments.
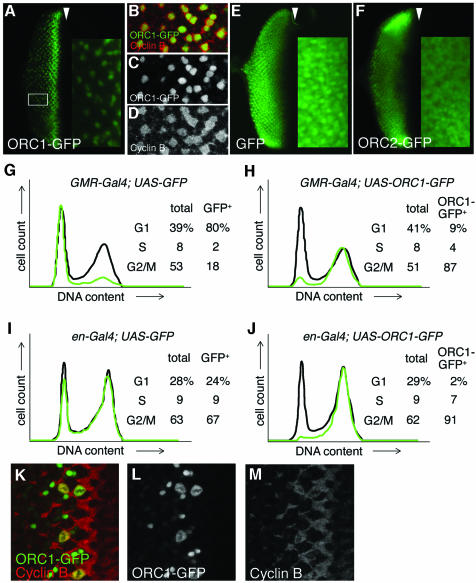
To our surprise, the accumulation of ORC1–GFP is essentially the same whether transcription is driven transiently by the orc1 promoter (Figure 1) or constitutively by the GMR promoter (Figure 3A). Protein levels first rise and then fall during the synchronous cycle in the eye disc as many of the cells go through mitosis. The majority of the cells are then in G1, with neither detectable CycB (not shown) nor ORC1–GFP, despite persistent transcription in the case of the ORC1–GFP transgene. A minor population of cells in the posterior region of the disc (e.g. boxed in Figure 3A) reside in a prolonged G2 arrest upon emerging from the morphogenetic furrow, since they bear high levels of CycB. Unlike the neighboring G1-arrested cells, these have persistent ORC1–GFP (Figure 3B–D). Taken together, these observations: (i) confirm the stability of ORC1–GFP in G2; and (ii) imply that cell cycle-mediated destruction of ORC1–GFP occurs not only at the M/G1 transition but continues into G1, even though the endogenous ORC1 substrate is normally present at negligible levels during this period due to inactivity of the ORC1 promoter. As a control, expression of GFP bearing a nuclear localization sequence (NLS) results in accumulation of protein to essentially the same level in all cells posterior to the furrow, regardless of their cell cycle phase (Figure 3E). Thus, cell cycle-dependent proteolysis of ORC1–GFP overrides constitutive transcription.
Fig. 3. Regulated proteolysis generates a normal temporal distribution of ORC1 protein even upon constitutive ORC1 transcription. Expression of various proteins under control of the GMR promoter (A–C, E–H, K and L), which is active in all cells posterior to the MF (arrowhead) in the eye disc, or the engrailed (en) promoter (I and J), which is active in all cells of the posterior compartment of the wing disc. High magnification views of the area of the eye disc boxed in (A) reveal that cells with high levels of ORC1–GFP are CycB positive with no BrdU incorporation (not shown), and therefore in G2 (B–D). FACS analysis reveals that ORC1–GFP is depleted in G1 phase eye (G and H) and wing (I and J) imaginal disc cells, even if transcription of ORC1–GFP is driven constitutively. High magnification views of cells immediately posterior to the MF reveal co-localization of ORC1–GFP and CycB following nuclear envelope breakdown and the presence of late telophase cells with paired nuclei bearing high levels of ORC1–GFP but no significant CycB (K–M).
The results of FACS analysis of eye antennal discs that constitutively express ORC1–GFP under GMR control support the view that ORC1 is degraded from the M/G1 boundary throughout G1. The stable control protein, GFP, is found primarily in the G1 cells that predominate in the region of the disc where the GMR promoter is active (Figure 3G). Despite this preponderance of G1 cells, ORC1–GFP is found almost entirely in the minor population of G2/M cells (Figure 3H). Because the eye disc cells posterior to the morphogenetic furrow constitute a developmentally unusual population, we wished to ascertain the behavior of ORC1–GFP upon constitutive expression in a more typical proliferating population of imaginal disc cells. To this end, we drove expression of ORC1–GFP (and, as a control, GFP) with the engrailed (en) promoter, which is constitutively active in cells of the posterior compartment during the third larval instar. As shown in Figure 3J, ORC1–GFP is essentially absent from G1 cells in these discs, whereas GFP is stable throughout the cell cycle (Figure 3I). Thus, the instability of ORC1–GFP during G1 appears to be a general property of imaginal disc cells.
Several additional experiments demonstrate that the behavior of ORC1–GFP is exceptional. First, ORC2–GFP behaves like the GFP control protein when driven by the GMR promoter, accumulating uniformly in every cell posterior to the furrow (Figure 3F). Secondly, constitutive transcription from transgenes encoding other cell cycle-regulated proteins is not masked by proteolysis. In particular, expression of CycE–GFP, E2F–GFP (not shown) or untagged versions of the same proteins (Richardson et al., 1995; Du et al., 1996) under GMR transcriptional control results in uniform protein accumulation throughout the posterior of the eye disc. In summary, the system responsible for degrading endogenous ORC1 at the end of M phase apparently is vigorous, active throughout G1 and relatively specific for ORC1.
When expressed under GMR promoter control, the ORC1–GFP fusion is detectable until somewhat later in M phase than is endogenous ORC1. As shown in Figure 3K–M, early in M phase ORC1–GFP co-localizes with CycB off the chromatin following nuclear envelope breakdown. Subsequently, CycB drops beneath the level of detection but ORC1–GFP appears to reassociate briefly with the DNA when it forms pairs of tightly condensed spheres that presumably correspond to late telophase nuclei. This residual ORC1–GFP is then abruptly degraded as the cells divide and enter G1. Note that accumulation of ORC1–GFP in presumptive telophase nuclei is also apparent when transcription is driven by the ORC1 promoter (arrows in Figure 1H). We do not know whether this minor difference in the behavior of endogenous ORC1 and ORC1–GFP is due to our ability to detect lower levels of ORC1–GFP, to enhanced transcription or to (modestly) enhanced stability of ORC1 by attachment of the GFP moiety. In any case, by using GMR to drive its expression, we find that ORC1–GFP levels fall precipitously upon exit from mitosis and that protein newly synthesized in G1 is also rapidly degraded and thus fails to accumulate to an appreciable extent.
Cell cycle-dependent degradation mediated by N-terminal signals
ORC1 is a member of a superfamily of ATP-binding proteins involved in DNA replication control that also includes ORC4, ORC5 and CDC6 (Bell et al., 1995; Perkins and Diffley, 1998; Tugal et al., 1998; Neuwald et al., 1999). Homology among these proteins is restricted to the C-terminal portion of ORC1, which bears the Walker A and B motifs (Neufeld et al., 1998; Schepers and Diffley, 2001). The role of the the N-terminal ORC1 domain is not known for any experimental system, although, in the case of human ORC1, it has been suggested that the N-terminal domain harbors signals that mediate its degradation during S phase (Mendez et al., 2002). Because Drosophila ORC1 is degraded at a different time in the cell cycle, it was of interest to determine whether the relevant signals also reside in the N-terminal region of the protein or whether they are embedded in the conserved C-terminal domain.
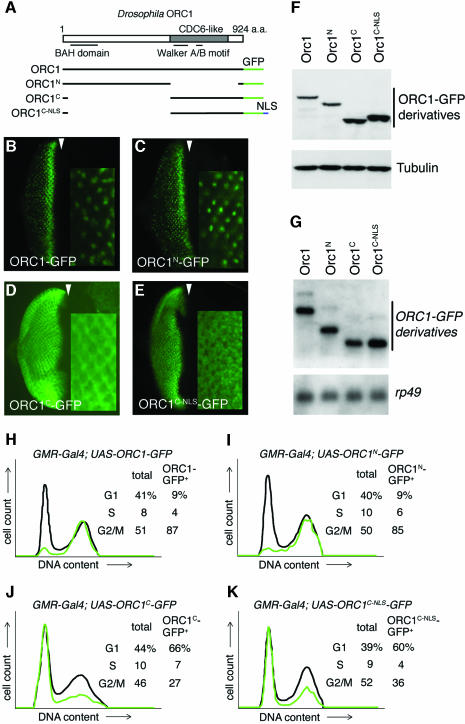
To map the degradation signals in Drosophila ORC1, transgenes that encode either the N- or C-terminal domain of ORC1 fused to GFP were prepared and transcription was driven in eye imaginal discs using the GMR promoter (described above). The stability of these derivatives was monitored both by examination of GFP fluorescence in situ and by FACS analysis of dissociated disc cells. As shown in Figure 4B, C, H and I, both assays reveal that ORC1N is regulated in a manner essentially indistinguishable from the full-length protein, degraded at the M/G1 boundary as well as throughout G1. In contrast, ORC1C is stable throughout the cell cycle, although it is predominantly cytoplasmic, presumably because it lacks a functional NLS (Figure 4D and J). To rule out the possibility that ORC1C is protected by virtue of its nuclear exclusion, we appended sequences that encode the SV40 NLS to the appropriate transgene and monitored accumulation of the encoded protein throughout the cell cycle, as described above. ORC1C–NLS is stable throughout the cell cycle when targeted to the nucleus (Figure 4E and K). Analysis of the level of protein (Figure 4F) and mRNA (Figure 4G) for each ORC1–GFP derivative supports the idea that the difference in steady-state accumulation of proteins bearing the ORC1 N-terminus is generated post-transcriptionally.
Fig. 4. Signals that mediate regulated proteolysis of ORC1 reside in its non-conserved N-terminal domain. (A) Schema of the deletion derivatives analyzed in (B–K). Expression of various ORC1 derivatives in the eye disc under GMR promoter control (B–E, each with a high magnification view inset), also analyzed by western blot (F), northern blot (G) and dissociation into single cells followed by FACS (H–K). The gels in (F) and (G) were re-probed with the loading controls shown at the bottom of each blot. Analysis of western blots reveals that the steady-state level of ORC1 or ORC1N is ∼25% that of ORC1C. Since these samples are homogenates of cells with low and high levels of protein, it underestimates the extent of regulation per cell (probably by a factor of ∼2, based on the cell cycle distributions of H–K).
In summary, the signals that mediate degradation of both human and Drosophila ORC1 appear to reside in the N-terminal domain of each protein, even though they are degraded at different stages of the cell cycle.
ORC1 degradation by the APC
In cultured cells, human ORC1 is degraded during S phase by Skp2-dependent SCF activity (Mendez et al., 2002). In contrast, the timing of ORC1 degradation in Drosophila strongly implies degradation by the APC, which degrades mitotic cyclins and securin to promote passage through and exit from mitosis. The APC is generally thought to be activated in succession, first by Fizzy (Fzy)/Cdc20, which promotes passage out of metaphase, and subsequently by Fizzy-related (Fzr)/Cdh1, which promotes exit from mitosis and suppresses CycB accumulation into G1. The ultimate consequence of APC activity is proteasome-dependent degradation of targeted substrates. The degradation of ectopically expressed ORC1 in G1 (Figure 3) suggests the involvement of Fzr.
To examine the role of Fzr, we wished to examine the behavior of ORC1 in mutant animals lacking Fzr activity. fzr mutants die in late embryogenesis, long before the imaginal discs can be studied; it also seems unlikely that fzr mutant somatic clones would proliferate and survive, precluding analysis in mosaic imaginal discs. Therefore, we analyzed epithelial cells in stage 12–13 embryos as they exit from M phase of division cycle 16 into G1 of cycle 17.
We first determined whether ORC1 and ORC1–GFP behave in a similar manner in these embryonic cells as in proliferating imaginal disc cells. In wild-type stage 12–13 embryos, most of the dorsal epithelial cells have entered a prolonged G1 arrest, with only a few cells still proliferating (Edgar and O’Farrell, 1990). These laggards have both detectable ORC1 and CycB, and thus are in S or G2 of cycle 16 (Figure 5A–C), whereas most of their neighbours have entered the prolonged G1 of cycle 17 and have neither protein. When expressed under the transcriptional control of the strong, constitutive actin5C promoter, ORC1–GFP accumulates to high levels only in the few epithelial cells still in S or G2, whereas the protein is destabilized in the remaining (G1) cells (Figure 5E). The selective degradation of ORC1–GFP in G1 is mediated by N-terminal signals, as is the case in imaginal disc cells (Figure 5F–H). Analysis of western and northern blots of embryonic samples (Figure 5I–J) is consistent with the idea that regulation of ORC1 accumulation is post-transcriptional. Thus, the stability of both the endogenous ORC1 and ectopically expressed ORC1–GFP is regulated in essentially the same fashion in the embryo and the imaginal disc.
Fig. 5. APC-dependent degradation of ORC1 in embryonic cells. Confocal images of epithelial cells just ventral to the leading edge in stage 12–13 embryos (A–H). At this stage, most cells are already in G1 of cycle 17 and only a minority of cells are in S or G2 of cycle 16, with high levels of both ORC1 and CycB (A–C). In (D–H), transcription of various proteins (as indicated) is driven by the constitutive Actin5C promoter. The few cells with high levels of ORC1–GFP or ORC1N–GFP also contain appreciable CycB and therefore are still in G2 of cycle 16 (not shown). We used northern blots (J) to select transgenic UAS lines that direct transcription of essentially identical levels of each gene, and then analyzed the steady-state level of each ORC1–GFP derivative by western blot (I). This analysis reveals that the steady-state level of ORC1C and ORC1C–NLS is 4- to 6-fold higher than ORC1–GFP and 6- to 9-fold higher than ORC1N. As in Figure 4, this analysis underestimates the extent of regulation per cell. Transmitted light microscopy reveals accumulation of CycB (K versus L) and ORC1 (M–P) in essentially every epithelial cell of fzr mutant embryos. Note that differential accumulation in internal tissues (primarily midgut) partially obscures accumulation in epithelial cells.
Next, we examined the dependence of ORC1 and CycB degradation on Fzr, comparing the accumulation of each protein in sibling wild-type and fzr mutant embryos. Essentially every epithelial cell in fzr mutant embryos has both appreciable ORC1 and CycB (Figure 5K–P; see also Jacobs et al., 2002). This observation is consistent with the idea that ORC1 degradation is dependent on Fzr. However, loss of Fzr activity perturbs the cell cycle, promoting epithelial cells into an extra division (Sigrist and Lehner, 1997). The accumulation of ORC1 might simply correlate with the progression of these cells into S and G2, where we have shown ORC1 is stabilized.
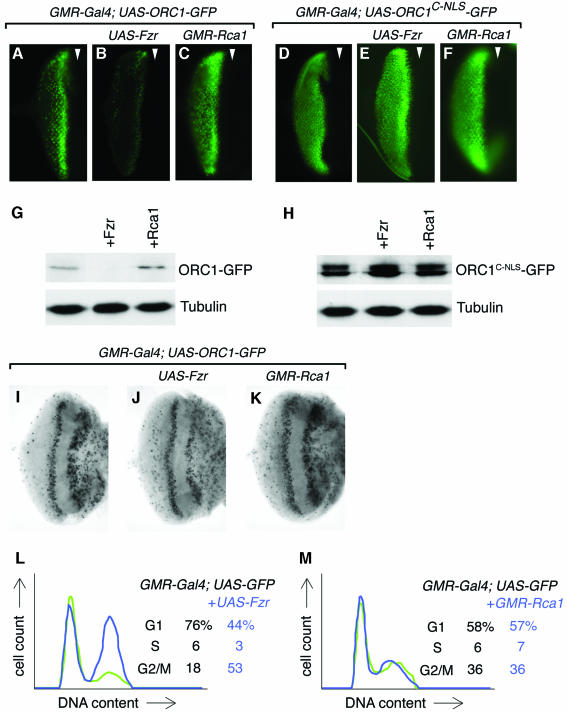
To better test the role of Fzr in regulating ORC1 stability in vivo, we therefore wished to perturb its activity without causing attendant dramatic changes in the cell cycle profile. To this end, we co-expressed either ORC1–GFP or the stable ORC1C–NLS–GFP derivative (as a control) in the eye imaginal disc with either Fzr or Rca1, a specific inhibitor of Fzr (Grosskortenhaus and Sprenger, 2002). Overexpression of Fzr causes a dramatic destabilization of ORC1–GFP, but not ORC1C–NLS–GFP (Figure 6A–H). Conversely, co-expression of Rca1 modestly stabilizes ORC1–GFP but has no effect on ORC1C–NLS–GFP. Importantly, the effect of ectopic Fzr and Rca1 cannot readily be explained by primary effects on cell cycle progression with subordinate effects on ORC1 stability. Misexpression of neither Fzr nor Rca1 promotes significant S phase entry in the eye disc cells (Figure 6I–K). Misexpression of Rca1 has no apparent effect on cell cycle progression analyzed by FACS (Figure 6M), and misexpression of Fzr actually leads to accumulation of additional cells in G2 (where ORC1 is normally stable) (Figure 6L). In similar experiments, we found that co-expression of Fzy had no apparent effect on ORC1–GFP stability (not shown), suggesting that ORC1 is preferentially targeted by Fzr. The simplest interpretation of these findings is that ORC1 is normally degraded upon exit from mitosis by Fzr-dependent APC activity.
Fig. 6. APC-mediated degradation of ORC1–GFP in imaginal cells. Co-expression of Fzr, which stimulates APC-dependent substrate degradation, significantly destabilizes ORC1–GPF in the eye disc (B versus A), whereas co-expression of Rca1, an inhibitor of Fzr, has the opposite effect (C). No effect was seen on the stability of the unregulated C-terminal domain of ORC1 (D–F). Western blots of the same disc samples are shown in (G) and (H). The distribution of BrdU incorporated during a brief pulse reveals that misexpression of neither Fzr nor Rca1 drives cells ectopically into S phase (I–K), although we see a slight effect similar to that reported (Dong et al., 1997) in the latter case. FACS analysis (L and M) reveals that misexpression of Fzr causes a modest accumulation of G2 cells (at the expense of the G1 population), whereas misexpression of Rca1 has no effect on the profile of cells in the posterior of the eye disc where the GMR promoter is active. Note that these experiments were performed in the absence of UAS-ORC1–GFP transgenes, but that expression of ORC1–GFP derivatives alone does not alter cycling of eye disc cells (Figures 2–4).
To test the idea that Fzr acts directly to target ORC1 for degradation, we asked whether Drosophila ORC1 is an APC substrate in vitro using a heterologous purified system (Fang et al., 1998). As shown in Figure 7A, ubiquitylation of a positive control, human polo-like kinase (Plk1), is stimulated by addition of APC activators from humans and flies, including Drosophila Fzr. Ubiquitinylation of ORC1N is also stimulated by Drosophila Fzr (and the human homolog, Cdh1; Figure 7B), whereas none of the APC activators tested significantly stimulates ubiquitylation of ORC1C (Figure 7C). These observations strongly support the idea that Drosophila ORC1 is targeted for degradation by Fzr by a mechanism that is conserved between vertebrates and flies.
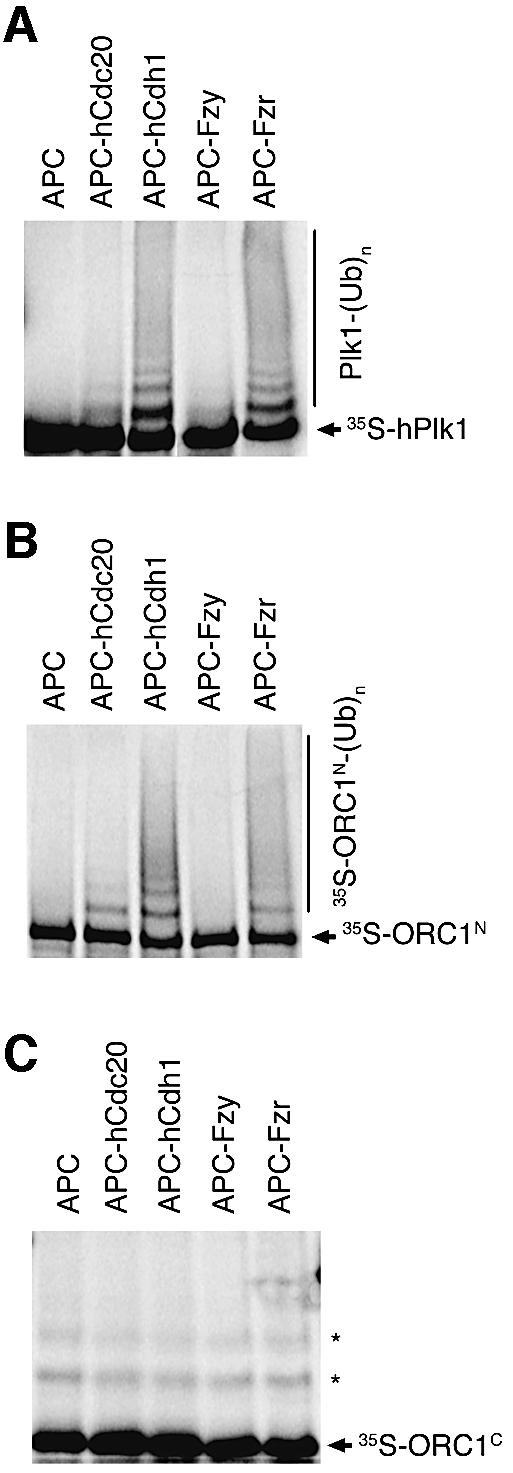
Fig. 7. Ubiquitylation of ORC1N by purified APC in vitro. Human Plk1 (A), Drosophila ORC1N (B) and Drosophila ORC1C (C) were incubated with purified Xenopus APC alone (lane 1 of each panel), or APC supplemented with the activator indicated above each lane. Ubiquitylation was detected by the generation of a higher molecular weight ladder following electrophoresis and autoradiography. The asterisks in (C) mark non-specifically ubiquitylated forms of the ORC1C substrate independent of active APC.
Discussion
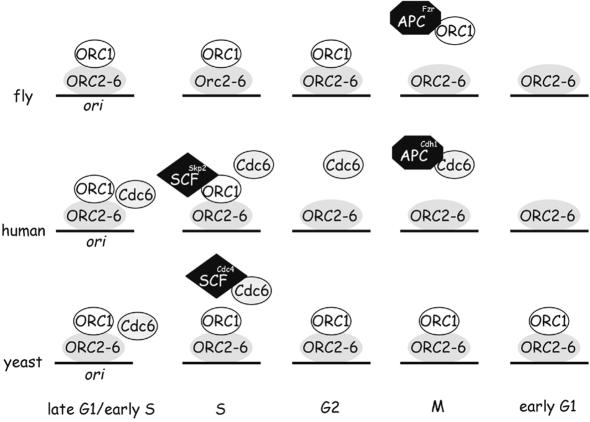
Replication origin activity in a variety of organisms is regulated by Cdc6 recruitment (for reviews see Bogan et al., 2000; Kelly and Brown, 2000; Bell and Dutta, 2002). The experiments described here suggest that modulation of ORC1 levels during cell cycle progression may provide an additional mechanism for controlling replication origin use in metazoans. Unlike ORC1 in budding and fission yeasts, human and fly ORC1 is available during only part of the cell cycle. Thus, ORC1 recruitment to the origin may fulfill some or all of the function supplied by Cdc6 recruitment in Saccharomyces cerevisiae, transforming an inert DNA-bound apo- complex to a replication-competent complex (Figure 8). According to this idea, regulated degradation of ORC1 is intrinsic to cell cycle progression in human and fly cells, allowing both organisms readily to regulate origin usage during development via ORC1. While this idea is generally attractive (Cimbora and Groudine, 2001; Li and DePamphilis, 2002; Mendez et al., 2002), to date the only direct supporting evidence comes from flies, where origin utilization in at least two different cell types is sensitive to the level of ORC1 (Asano and Wharton, 1999).
Fig. 8. Model of our current understanding of the temporal distribution of ORC1 in fly, human and either budding or fission yeast cells. Drosophila Cdc6 is uncharacterized and therefore omitted.
Different aspects of cell cycle progression are driven by two proteolytic machines: the SCF ubiquitin ligase family and the APC. In principle, either proteolytic system could suffice to degrade ‘spent’ ORC1 after the initiation of S phase, thereby helping to reset origins and prevent re-replication before completion of the cycle. Why then do human and fly cells degrade ORC1 at different stages of the cell cycle using different mechanisms? Three possible explanations are discussed below.
One possible explanation is that degradation via the APC is a mechanism to restrict origin licensing to a very narrow window at the end of M or the onset of G1. As shown in Figure 3K–M, there is a brief period during the M/G1 transition when CycB is absent but ORC1–GFP is not yet degraded. Licensing might occur only during this interval, with subsequent Fzr-dependent degradation of ORC1 limiting origin activation to the beginning of G1. Note that we cannot exclude the possibility that ORC1 acts late in G1 to license origins. In either case, the timing of origin activation may be different in flies and vertebrates, where licensing is generally permitted throughout much of G1 (for a review see Bell and Dutta, 2002).
Another possible explanation is that evading degradation at the M/G1 boundary may facilitate the switch from a canonical four-phase cell cycle to an endocycle. Throughout the life of the fly, many cells bypass M phase and re-replicate their DNA. These cells might be able to reuse ORC1 from one cycle to the next by bypassing that portion of the cell cycle when the APC is active, from M phase into G1. In contrast, if fly ORC1 were degraded shortly after the onset of DNA synthesis, as is the case for human ORC1, the next round of DNA synthesis would require de novo synthesis of ORC1. Such a strategy would be less efficient; it might also be impossible in differentiated cells in which E2F transcription factor activity, required for ORC1 transcription in cycling cells, is inhibited or absent (for a review see Dyson, 1998).
Another possible explanation is that the ORC may have important biological roles outside S phase, in which case prolonging the life of ORC1, the least stable component, through a greater portion of the cell cycle might be important. Three lines of evidence support the idea that the Drosophila ORC is involved in events outside S phase. First, genetic studies reveal that the ORC appears to play a role in regulating chromosome condensation and position effect variegation (Pak et al., 1997; Loupart et al., 2000). ORC1 may play a central role in mediating such events, as it interacts with the heterochromatin-associated protein HP-1 (Pak et al., 1997). Secondly, the terminal phenotype of cells in ORC2 and ORC5 mutant flies is not S phase arrest, as might be expected, but rather M phase arrest with irregular chromosome condensation (although these phenotypes might be secondary to defects in the rate of DNA synthesis) (Loupart et al., 2000; Chesnokov et al., 2001). Thirdly, the phenotype of ORC3 mutant flies, as well as the distribution of ORC3 protein, suggest a novel role in controlling synaptic plasticity that is independent of the cell cycle (Pinto et al., 1999; Rohrbough et al., 1999). Further definition of the role of Drosophila ORC1 will require characterization of an appropriate mutant.
The N-terminal domain of ORC1 contains a number of signals that mediate APC-dependent degradation of other proteins, including a KEN box, several canonical D boxes (RxxLxxxxN/Q/E/D), as well as a variant D box (KxxLxxxxN) that has been shown to mediate APC-dependent degradation of Drosophila securin (Leismann and Lehner, 2003). Our preliminary analysis suggests that none of these is responsible for the regulated degradation of ORC1 (not shown). This observation is not terribly surprising, given the recent identification of a number of other signals that mediate APC-dependent degradation of substrates in other systems (Castro et al., 2002, 2003; Littlepage and Ruderman, 2002). The Drosophila ORC1 signal apparently is recognized by vertebrate factors (Figure 7), and thus it will be of interest in future to define the signal and determine whether it mediates destruction of other fly and mammalian proteins.
Although transcriptional and post-translational mechanisms appear to contribute to setting the ORC1 level in Drosophila, cell cycle-regulated degradation plays the dominant role. E2F-dependent transcription of the ORC1 gene is narrowly confined to late G1 and early S phase, yielding a relatively brief window for the generation of protein (Asano and Wharton, 1999). Although constitutive proteolysis might suffice to degrade this protein within a relatively brief window, in fact ORC1 degradation is itself cell cycle regulated to allow catastrophic disappearance of ORC1 abruptly during exit from mitosis. Moreover, persistent APC activity into G1 is sufficient to generate an essentially normal temporal distribution of ORC1 protein even if transcription of ORC1 is uncoupled from its normal, E2F-dependent signals and driven constitutively. We imagine that, in the absence of such potent proteolysis, transcriptional overexpression of ORC1 would cause much more striking organismal and cellular phenotypes than have been reported.
The work reported here extends the structural and functional analogy between ORC1 and Cdc6 described previously. Both ORC1 (in flies) and Cdc6 (in the yeasts) can govern origin utilization. Also, both ORC1 (in flies) and Cdc6 (in mammalian cells) are degraded by the APC. Thus, ORC1 appears to serve dual functions at origins of replication, acting as both a regulatory factor and a structural component of the origin-binding complex.
Materials and methods
Strains and reagents
Mouse and rabbit anti-CycB antibodies and fzrie28, UAS-Fzr lines, the GMR-Rca1 line, affinity-purified anti-GFP rabbit IgG, and mGFP6 plasmid DNA were generously provided by C.Lehner, B.Thomas, P.Silver and J. Raff, respectively. Monoclonal DM1A anti-tubulin antibodies were from Sigma. GMR-Gal4, UAS–GFP-NLS, en-Gal4 and Actin5-Gal4 lines were obtained from Bloomington stock center. Other transgenic flies were generated by microinjection of w1118 embryos by standard methods.
Plasmid construction
ORC1–GFP fusion expression plasmid was constructed by replacing the termination codon of ORC1 cDNA with an Asp718 site to fuse mGFP6 cDNA (Schuldt et al., 1998) with six copies of a 13 amino acid peptide recognized by mAb 9E10 in-frame. The 2.4 kbp orc1 promoter (Asano and Wharton, 1999), ORC1–GFP cDNA and poly(A) signal provided by the α-tublin 3′-untranslated region (UTR) were inserted into the pCaSpeR vector for P-element transformation. To delete the N- or C-terminal portion of ORC1, the SgrAI site was fused to the NheI site or the NheI site was fused to the EcoRIII site, respectively. Each ORC1–GFP fusion cDNA was inserted into the pUASP vector (Rorth, 1998) with the α-tublin 3′-UTR for P-element transformation.
Histology
Third instar larval eye discs were dissected in phosphate-buffered saline (PBS; 130 mM NaCl, 7 mM Na2HPO4 and 3 mM NaH2PO4), fixed with 2% paraformaldehyde (PFA) in PBS for 30 min, blocked and permeabilized in PGS (1% goat serum and 0.1% saponin in PBS) for 30 min at room temperature. Samples were reacted with primary antibodies in PGS at 4°C overnight. Secondary antibodies coupled to fluorescein isothiocyanate (FITC) or Texas red (Jackson Laboratories) were used for visualization. In Figure 1C, E and F, discs were fixed for 10 min, blocked and permeabilized in PGST (3% goat serum, 0.2% saponin and 0.2% Triton X-100 in PBS), and reacted with primary antibodies in PGT (1% goat serum, 0.2% Triton X-100 in PBS). Alexa 488-conjugated anti-FITC (Molecular Probes) was used as tertiary antibody. Primary antibodies were used at the following concentrations; 1:1000 rabbit anti-CycB; 1:100 affinity-purified rat anti-ORC1; 1:6000 rabbit anti-PH3 (Upstate); and 1:3000 mouse anti-myc (9E10, Santa Cruz).
For double staining experiments of ORC1 and CycB (Figure 5A–C), embryos were fixed as described previously (Asano and Wharton, 1999), blocked with TBTG [1% goat serum, 1% bovine serum albumin (BSA), 0.1% Triton X-100, 0.25 M NaCl and 10 mM Tris–HCl pH 8.0] and reacted with 1:100 rat anti-ORC1 and 1:500 rabbit anti-CycB in TBTG at 4°C overnight. FITC-conjugated anti-rat IgG and Texas red-conjugated anti-rabbit IgG were used as secondary antibodies. The embryos in Figure 5D–H and K–P were dechorionated, fixed in 4% PFA in PBS under heptane, devitellinized into methanol, and rehydrated into PBS. A mixture of sibling fzr mutant and wild-type embryos (Figure 5K–P), obtained from a fzrie28/FM7C, P[w+, fts-lacZ] stock (Jacobs et al., 2002), was blocked with PBTG (1% goat serum and 0.1% Triton X-100 in PBS) and double-stained with 1:100 rabbit anti-LacZ (Cappel) and 1:400 rat anti-ORC1 or 1:3 mouse anti-Cyc B (F2F4) in PBTG at 4°C overnight. ORC1 and CycB signals were first visualized by 3′,3′-diaminobenzidine (DAB) staining using horseradish peroxidase (HRP)-conjugated secondary antibodies. Embryos were then sorted by LacZ expression visualized by an FITC-conjugated secondary antibody. Bromodeoxyuridine (BrdU) labeling was performed as described previously (Asano et al., 1996), except Schinerder’s medium (Gibco) was used in place of Ringer’s solution. The dissociation of imaginal disc cells and FACS analysis were performed following the protocol from the Edgar laboratory (Neufeld et al., 1998).
In vitro ubiquitylation
Assays were carried out essentially as described previously (Fang et al., 1998), using 35S-labeled substrates generated by coupled transcription/translation in vitro, supplementing the APC with activators also generated by coupled transcription/translation in vitro.
Acknowledgments
Acknowledgements
We particularly thank Christian F.Lehner for his exceptional generosity, as well as Barbara J.Thomas, Andrea H.Brand, Jordan W.Raff and Pamela A.Silver for reagents, Mami Kawaguchi, Cary D.Gardner and Tammy Lee for technical help, Glenda Johnson and Jian Chen for fly media preparation, Sandy Boyles for secretarial help, Mike J.Cook and Lynn M.Martinek for FACS analysis, Robert J.Duronio, Danny J.Lew and Joseph R.Nevins for critical comments on the manuscript, and an anonymous reviewer for pointing out the implications for origin licensing. We are grateful to J.R.Nevins for his generous support, encouragement and extraordinary patience throughout this work. M.A. is supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science. R.P.W. is an Associate Investigator of the HHMI. This work was supported by National Institutes of Health grants to M.A. (GM64348) and to H.Y. (GM61542).
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S.cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Asano M., Nevins,J.R. and Wharton,R.P. (1996) Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev., 10, 1422–1432. [DOI] [PubMed] [Google Scholar]
- Asano M. and Wharton,R.P. (1999) E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J., 18, 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. (1995) Eukaryotic replicators and associated protein complexes. Curr. Opin. Genet. Dev., 5, 162–167. [DOI] [PubMed] [Google Scholar]
- Bell S.P. (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev., 16, 659–672. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Mitchell,J., Leber,J., Kobayashi,R. and Stillman,B. (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell, 83, 563–568. [DOI] [PubMed] [Google Scholar]
- Bogan J.A., Natale,D.A. and Depamphilis,M.L. (2000) Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol., 184, 139–150. [DOI] [PubMed] [Google Scholar]
- Castro A., Vigneron,S., Bernis,C., Labbe,J.C., Prigent,C. and Lorca,T. (2002) The D-box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-box sequence of Aurora-A. EMBO Rep., 3, 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A., Vigneron,S., Bernis,C., Labbe,J.C. and Lorca,T. (2003) Xkid is degraded in a D-box, KEN-box and A-box-independent pathway. Mol. Cell. Biol., 23, 4126–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I., Remus,D. and Botchan,M. (2001) Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl Acad. Sci. USA, 98, 11997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimbora D.M. and Groudine,M. (2001) The control of mammalian DNA replication: a brief history of space and timing. Cell, 104, 643–646. [PubMed] [Google Scholar]
- Datar S.A., Jacobs,H.W., de La Cruz,A.F., Lehner,C.F. and Edgar,B.A. (2000) The Drosophila cyclin D–Cdk4 complex promotes cellular growth. EMBO J., 19, 4543–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. (2001) DNA replication: building the perfect switch. Curr. Biol., 11, R367–R370. [DOI] [PubMed] [Google Scholar]
- Dong X., Zavitz,K.H., Thomas,B.J., Lin,M., Campbell,S. and Zipursky,S.L. (1997) Control of G1 in the developing Drosophila eye: rca1 regulates cyclin A. Genes Dev., 11, 94–105. [DOI] [PubMed] [Google Scholar]
- Du W., Xie,J.-E. and Dyson,N. (1996) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J., 15, 3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. and O’Farrell,P.H. (1990) The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell, 62, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Findeisen M., El-Denary,M., Kapitza,T., Graf,R. and Strausfeld,U. (1999) Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6 and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem., 264, 415–426. [DOI] [PubMed] [Google Scholar]
- Fujita M., Ishimi,Y., Nakamura,H., Kiyono,T. and Tsurumi,T. (2002) Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem., 277, 10354–10361. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R. and Sprenger,F. (2002) Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell, 2, 29–40. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Burton,J.L. and Solomon,M.J. (2002) The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev., 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Hua X.H. and Newport,J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol., 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. and Raff,J.W. (1999) The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J., 18, 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H., Richter,D., Venkatesh,T. and Lehner,C. (2002) Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr. Biol., 12, 1435–1441. [DOI] [PubMed] [Google Scholar]
- Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- Lehner C.F. and O’Farrell,P.H. (1990) The roles of Drosophila cyclins A and B in mitotic control. Cell, 61, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leismann O. and Lehner,C.F. (2003) Drosophila securin destruction involves a D-box and a KEN-box and promotes anaphase in parallel with cyclin A degradation. J. Cell Sci., 116, 2453–2460. [DOI] [PubMed] [Google Scholar]
- Li C.J. and DePamphilis,M.L. (2002) Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol., 22, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage L.E. and Ruderman,J.V. (2002) Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev., 16, 2274–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupart M.L., Krause,S.A. and Heck,M.S. (2000) Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr. Biol., 10, 1547–1556. [DOI] [PubMed] [Google Scholar]
- Mendez J., Zou-Yang,X.H., Kim,S.Y., Hidaka,M., Tansey,W.P. and Stillman,B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell, 9, 481–491. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nature Cell Biol., 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- Moses K. and Rubin,G.M. (1991) Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev., 5, 583–593. [DOI] [PubMed] [Google Scholar]
- Natale D.A., Li,C.J., Sun,W.H. and DePamphilis,M.L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M–G(1) transition in mammals. EMBO J., 19, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz,A.F., Johnston,L.A. and Edgar,B.A. (1998) Coordination of growth and cell division in the Drosophila wing. Cell, 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Newlon C.S. and Theis,J.F. (1993) The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev., 3, 752–758. [DOI] [PubMed] [Google Scholar]
- Ohtani K., James,D., Leone,G., Herendeen,D.R., Kelly,T.J. and Nevins,J.R. (1996) Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol., 16, 6977–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D.T., Pflumm,M., Chesnokov,I., Huang,D.W., Kellum,R., Marr,J., Romanowski,P. and Botchan,M.R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Perkins G. and Diffley,J.F. (1998) Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell, 2, 23–32. [DOI] [PubMed] [Google Scholar]
- Pinto S. et al. (1999) latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron, 23, 45–54. [DOI] [PubMed] [Google Scholar]
- Richardson H., O’Keefe,L.V., Marty,T. and Saint,R. (1995) Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development, 121, 3371–3379. [DOI] [PubMed] [Google Scholar]
- Ritzi M., Baack,M., Musahl,C., Romanowski,P., Laskey,R.A. and Knippers,R. (1998) Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem., 273, 24543–24549. [DOI] [PubMed] [Google Scholar]
- Rohrbough J., Pinto,S., Mihalek,R.M., Tully,T. and Broadie,K. (1999) latheo, a Drosophila gene involved in learning, regulates functional synaptic plasticity. Neuron, 23, 55–70. [DOI] [PubMed] [Google Scholar]
- Rorth P. (1998) Gal4 in the Drosophila female germline. Mech. Dev., 78, 113–118. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada,S. and Blow,J.J. (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci., 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Chen,J., Thome,K.C., Lawlis,S.J., Hou,Z.H., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A. and Diffley,J.F. (2001) Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J. Mol. Biol., 308, 597–608. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams,J.H., Davidson,C.M., Micklem,D.R., Haseloff,J., St Johnston,D. and Brand,A.H. (1998) Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. (1996) Cancer cell cycles. Science, 274, 1672–1677. [DOI] [PubMed] [Google Scholar]
- Sigrist S.J. and Lehner,C.F. (1997) Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell, 90, 671–681. [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs,H., Stratmann,R. and Lehner,C.F. (1995) Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J., 14, 4827–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. (1996) Cell cycle control of DNA replication. Science, 274, 1659–1664. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y., Tsurimoto,T., Shirahige,K., Yoshikawa,H. and Obuse,C. (2000) Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem., 275, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Tugal T., Zou-Yang,X.H., Gavin,K., Pappin,D., Canas,B., Kobayashi,R., Hunt,T. and Stillman,B. (1998) The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem., 273, 32421–32429. [DOI] [PubMed] [Google Scholar]
- Whitfield W.G., Gonzalez,C., Maldonado-Codina,G. and Glover,D.M. (1990) The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2–M transition. EMBO J., 9, 2563–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]