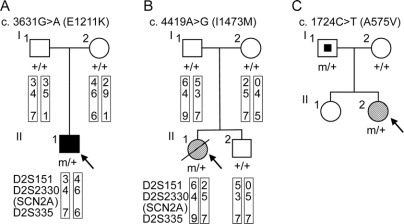
Figure 1 Novel SCN2A nucleotide changes leading to amino acid substitutions identified in patients with intractable childhood epilepsies
(A) Pedigree for proband 1 (II-1) with a history of infantile spasms that evolved to symptomatic generalized epilepsy who showed the nucleotide change c.3631C>T (E1211K). The putative haplotypes determined by analyzing microsatellite markers, namely, D2S151, D2S2330, and D2S335, flanking the SCN2A locus are shown. Markers are given in order, from the p telomere to the q telomere. + = Wild-type allele; m = mutated allele; filled square = infantile spasms. (B) Pedigree for proband 2 (II-1) with neonatal epileptic encephalopathy who showed the nucleotide change c.4419A>G (I1473M). The putative haplotypes determined by analyzing microsatellite markers flanking the SCN2A locus are shown. Genetic recombination occurred in the proband’s or her brother’s maternal chromosome within the region between D2S151 and D2S2330. + = Wild-type allele, m = mutated allele; hatched circle = neonatal epileptic encephalopathy; slash = deceased. (C) Pedigree for proband 3 (II-2) with SMEB who showed the nucleotide change c.1724C>T (A575V). The nucleotide change was also detected in her asymptomatic father. Genomic DNA of her sister was not available for the analysis. + = Wild-type allele; m = mutated allele; hatched circle = SMEB; symbol with a dot = asymptomatic mutation carrier.