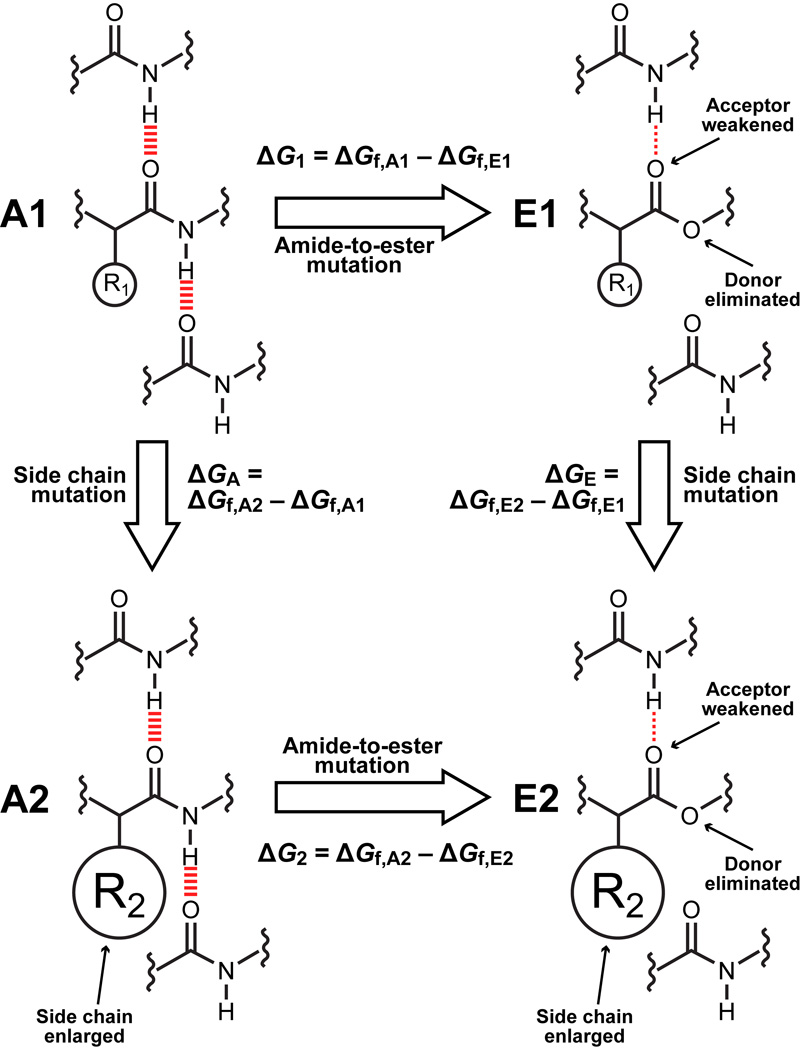
Figure 1. Double mutant cycle design.
Amide-to-ester mutation of the backbone perturbs hydrogen bonding while traditional side chain mutation of a nearby residue perturbs microenvironment polarity. The differences in the free energies of folding of the pairs of mutants A1/E1 and A2/E2, ΔG1 = ΔGf,A1 – ΔGf,E1 and ΔG2 = ΔGf,A2 – ΔGf,E2 respectively, primarily (but not exclusively) reflect the strength of the hydrogen bond lost in the context of a smaller, less hydrophobic side chain (A1/E1) or a larger, more hydrophobic side chain (A2/E2). The differences in the free energies of folding of the pairs of mutants A2/A1 and E2/E1, ΔGA = ΔGf,A2 – ΔGf,A1 and ΔGE = ΔGf,E2 – ΔGf,E1 respectively, primarily reflect the effect of the side chain on folding free energy in the presence or absence of the hydrogen bond(s) formed by the amide of interest. The thermodynamic coupling energy, ΔΔG2–1 = ΔG2 – ΔG1 = ΔGA – ΔGE reflects the effect of the microenvironment on hydrogen bond strength (barring a structural rearrangement in the amide-to-ester mutants): if it is negative, the hydrogen bond is stronger in the more hydrophobic microenvironment; if it is positive, the opposite is true; finally, if it is 0, then either the hydrogen bond is unaffected by its microenvironment or its microenvironment does not change upon folding.