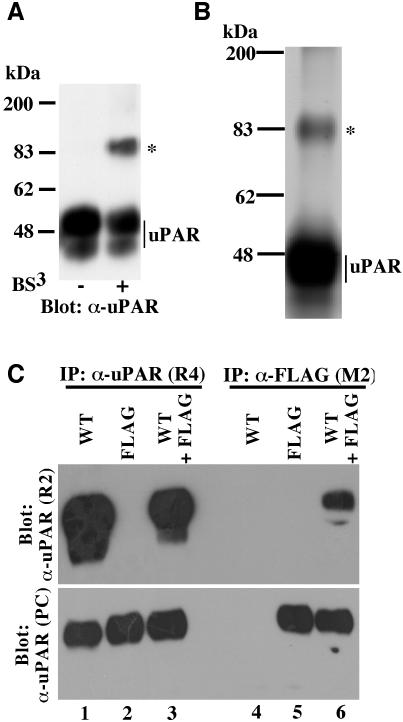
Fig. 2. Dimeric cell surface uPAR revealed by chemical cross linking and co-immunoprecipitation. (A) Western blot analysis of uPAR dimerization by chemical cross linking. 293/uPAR cells were treated with the chemical cross linker BS3 as indicated below the panel, washed and lysed. Equal amounts of total protein were separated by SDS–PAGE and analyzed by immunoblotting using a polyclonal anti-uPAR antibody. Similar data were observed in several experiments. (B) Affinity purification of cross-linked uPAR. Lysates obtained from large-scale cross linking experiments carried out on 293/uPAR cells were loaded onto an anti-uPAR antibody affinity column. After washing, bound protein was eluted, concentrated and separated by SDS–PAGE. The slab gel was silver stained and the band corresponding to the uPAR adduct (marked by an asterisk) excised, subjected to trypsin digestion and analyzed by mass spectrometry (see Supplementary data). (C) Lysates from 293 cells expressing either wild-type uPAR (WT), FLAG-tagged uPAR (FLAG) or both (WT+FLAG) were immunoprecipitated with an anti-uPAR antibody (R4) which recognizes both wild-type and FLAG-tagged uPAR, or with an anti-FLAG antibody (M2) which recognizes only the FLAG-tagged receptor. The immunoprecipitated material was fractionated by SDS–PAGE and analyzed by immunblotting using an anti-uPAR antibody (R2) which recognizes only wild-type uPAR (upper panel). To ensure that appropriate immunoprecipitation had been achieved, the blots were also probed with a polyclonal uPAR antibody which recognizes all forms of the receptor (α-uPAR-PC, lower panel). Similar results were obtained in at least three independent experiments.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.