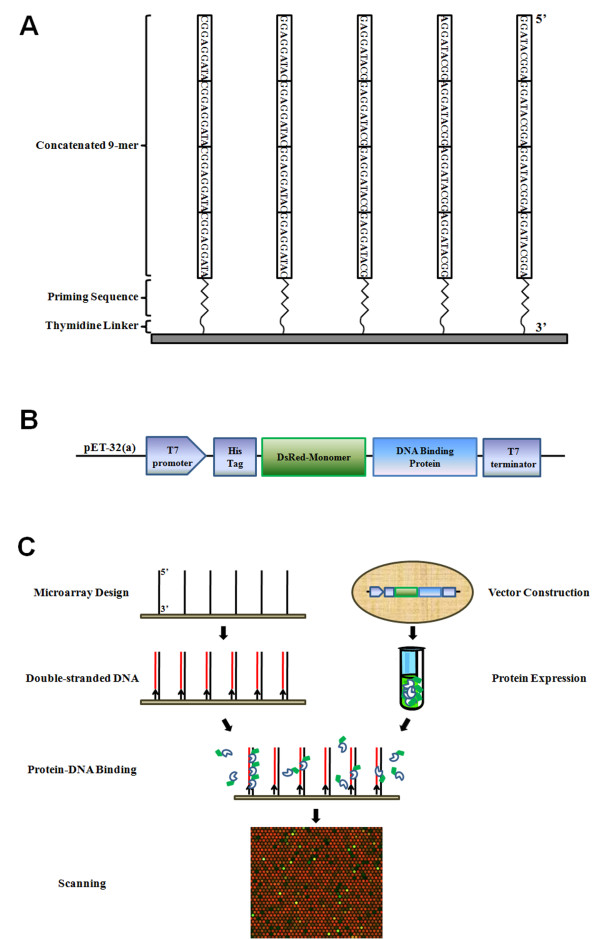
Figure 1.
Experiment using the Quadrupled 9-mer Protein Binding Microarray (Q9-PBM). (A) All possible combinations of 9-mer oligonucleotides were quadrupled, and then followed by a primer binding sequence and a 5-nt thymidine linker attached to the slide. A total of 232,145 probe features, including 131,072 features from all possible 9-mers and 101,073 replicated features, were designed. (B) The pET32-DsRed expression vector was constructed. Proteins were expressed with an N-terminal fusion to the polyhistidine-tag and red fluorescent protein (DsRed). The DsRed fluorescent protein was cloned from Discosoma sp. based on homology with the green fluorescent protein (GFP). DsRed possesses a similar spectrum to Cy3 that is compatible with the microarray scanner. The full length cDNA of the transcription factors was cloned into the pET32(a) expression vector followed by DsRed fluorescent protein. (C) The reverse complimentary strand was synthesized on a slide by thermo-stable DNA polymerase. A small quantity (1.6 μM) of Cy5 fluorescent dUTP was incorporated to confirm successful elongation. Purified DsRed fusion protein was incubated with the double-stranded microarray. The consensus sequence was determined from the fluorescence intensity of the spot without any further step like antibody labeling. The presented scanning image is the part of the Cbf1 result.