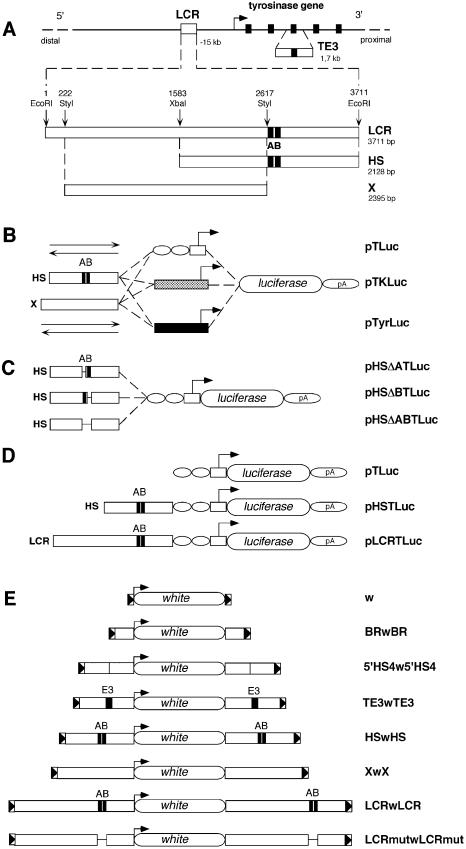
Figure 1.
DNA constructs used for in vitro and in vivo assays. (A) Schematic view of the mouse tyrosinase locus and LCR sequences in mouse chromosome 7. A bent arrow indicates the transcription start site and direction of the mouse tyrosinase gene. Exons (5) are indicated as black rectangles. Not drawn to scale. A 1.7 kb fragment containing mouse tyrosinase exon 3 (TE3) and surrounding intronic sequences is shown below the mouse tyrosinase gene. The 3.7 kb EcoRI fragment containing the mouse tyrosinase LCR, located at –15 kb of the gene is shown below (3711 bp, EMBL Database X76647). Relative position of the 3.7 kb EcoRI DNA fragment with respect to the start of transcription of the tyrosinase gene (–15 kb) has been updated, according to the latest available mouse genome sequence information (see legend for Fig. 5), therefore correcting the position assigned to this LCR fragment in previous reports (14,17). The relative position of restriction enzymes used to subclone internal fragments is indicated along with their size in base pairs (bp). A and B black boxes, referred to previously as HS-1 and HS-2 (14), identify binding sequences for nuclear factors. (B) Three series of backbone plasmids are shown: pTLuc, containing a TATA box from the HSV-TK gene (white box), linked to two sites for the ubiquitous transcription factor Sp1 (white ovals); pTKLuc, containing the promoter of the HSV-TK gene (grey box); and pTyrLuc, carrying the mouse tyrosinase promoter (black box). Upstream of each promoter, the HS and X fragments were cloned in either direction (depicted as two arrows) resulting in the plasmids pHSTLuc, pSHTluc, pXTLuc, pXinvTLuc, pHSTKLuc, pSHTKLuc, pXTKLuc, pXinvTKLuc, pHSTyrLuc, pSHTyrLuc, pXTyrLuc and pXinvTyrLuc, respectively. (C) Plasmids used for functional analysis of deletion mutants within the HS fragment. (D) Constructs used to evaluate boundary activities of mouse tyrosinase LCR sequences in transgenic mice. All constructs shown in (B), (C) and (D) share the firefly luciferase reporter gene and the 3′ splice site and polyadenylation signal from the small t gene of the SV40 genome, indicated as pA. (E) DNA constructs used to evaluate boundary activities of mouse tyrosinase LCR sequences in transgenic flies. ‘w’ represents the reference construct [pCaSpeR3 (51)] containing a white minigene surrounded by 3′P and 5′P sequences from a P element (shown as triangles in a box). BRwBR carries the Su(Hw) BR of gypsy retrotransposon (52), 5′HS4w5′HS4 carries two copies of the 1.2 kb fragment containing the 5′HS4 of the chicken β-globin locus (36), TE3wTE3, HSwHS, XwX, LCRwLCR and LCRmutwLCRmut contain the corresponding mouse tyrosinase LCR DNA fragments.