SUMMARY
Type IIS restriction endonuclease Eco31I is a ‘short-distance cutter’, which cleaves DNA strands close to its recognition sequence, 5′-GGTCTC(1/5). Previously, it has been proposed that related endonucleases recognizing a common sequence core GTCTC possess two active sites for cleavage of both strands in the DNA substrate. Here, we present bioinformatic identification and experimental evidence for a single nuclease active site. We identified a short region of homology between Eco31I and HNH nucleases, constructed a three-dimensional model of the putative catalytic domain and validated our predictions by random and site-specific mutagenesis. The restriction mechanism of Eco31I is suggested by analogy to the mechanisms of phage T4 endonuclease VII and homing endonuclease I-PpoI. We propose that residues D311 and N334 coordinate the cofactor. H312 acts as a general base activating water molecule for the nucleophilic attack. K337 together with R340 and D345 are located in close proximity to the active center and are essential for correct folding of catalytic motif, while D345 together with R264 and D273 could be directly involved in DNA binding. We also predict that the Eco31I catalytic domain contains a putative Zn-binding site, which is essential for its structural integrity. Our results suggest that the HNH-like active site is involved in the cleavage of both strands in the DNA substrate. On the other hand, analysis of site-specific mutants in the region, previously suggested to harbor the second active site, revealed its irrelevance to the nuclease activity. Thus, our data argue against the earlier prediction and indicate the presence of a single conserved active site in Type IIS restriction endonucleases that recognize common sequence core GTCTC.
Keywords: restriction endonuclease, Type IIS, HNH, endonuclease VII, active site
INTRODUCTION
Data on details of structural and functional organization of Type IIS restriction endonucleases (REases), which recognize asymmetric DNA sequences and cleave at least one DNA strand outside of the recognition sequence1, have been limited to FokI and BfiI. Proteolysis and later X-ray crystallography experiments demonstrated that FokI consists of two structural domains: an N-terminal domain, which recognizes and binds the DNA containing recognition sequence, whereas the C-terminal domain cleaves DNA non-specifically, and together they form an enzyme that cleaves the DNA at a specific distance from the recognition sequence.2,3 A monomer of FokI contains only one catalytic domain with the PD-(D/E)XK-like catalytic center, which dimerizes to cleave both strands of the DNA target.4 BfiI is unrelated to FokI, yet it also exhibits a two-domain structure. It comprises the N-terminal nuclease domain from the phospholipase D superfamily fused with the C-terminal DNA recognition domain.5 In contrast to FokI, BfiI is a dimer in solution and forms a single nuclease active site composed of residues from two catalytic domains.6 Recently, several Type IIS REases were reported to contain the third type of the nuclease domain, characterized by an HNH-like active site. An alignment of the C-terminal region of MnlI revealed homology to the HNH domain of bacterial DNases colicins and mutational analysis confirmed the importance of conserved residues for the nuclease activity of this enzyme.7 It was also found that the proteolytically separated C-terminal domain of MnlI cleaves DNA non-specifically, in analogy to the catalytic domain of FokI and BfiI.8 Sequence analyses suggested that another Type IIS enzyme, Hin4II, also belongs to the HNH nuclease superfamily.9 Bioinformatic and mutational analysis of Type IIS REase HphI have also provided evidence that this enzyme has a catalytic domain related to the HNH nucleases.10 The HNH motif was also indicated in KpnI, which belongs to Type IIP REases, the single domain enzymes. The analysis of KpnI mutants showed that HNH motif of KpnI is coupled with DNA recognition function, contrary to the HNH motifs indicated in Type IIS REases.11
Analysis of the reaction courses of various Type IIS REases revealed four modes of action. The group of Type IIS REases named as ‘short-distance cutters’, cleave plasmids containing one or two sites with similar rate. They act essentially like orthodox Type IIP enzymes and convert their substrates directly to the linear form without accumulating the nicked form of DNA. Two mechanisms for their action have been hypothesized: either these enzymes are monomers carrying two active sites or they are homodimeric proteins with one active site in each subunit.12 Recently, Mva1269I has been found to conform to the first model – this Type IIS enzyme possesses two PD-(D/E)XK domains, which cleave the DNA target sequentially.13 The same was found for BsmI (isoschizomer of Mva1269I) by Xu et al. (International Patent WO 200612593 A, 2006). Experiments aimed at producing nicking enzymes from ‘short-distance cutters’ were also reported for enzymes SapI, BsaI, BsmBI, and BsmAI, where the selection procedures were based on the two-center hypothesis and involved selective inactivation of one of the two putative catalytic domains.14,15 However, thus far no structural models of these enzymes were available to guide rational protein engineering experiments.
Here, we describe the identification of the HNH-like active center in Type IIS REase Eco31I, which belongs to ‘short-distance cutters’ and cleaves both strands of DNA close to the target site 5′-GGTCTC(1/5).16,17 Using bioinformatic methods we constructed a structural model of the catalytic domain and used it as a platform to interpret the old and new experimental data. We present evidence that Eco31I possesses a single active site that preferentially cleaves either a top or the bottom strand of the DNA substrate, depending on the sequence context. Therefore, we supposed that the same features are characteristic for the other related enzymes, which recognize common sequence core 5′-GTCTC, including Alw26I, BsaI, BsmAI, BsmBI and Esp3I.
RESULTS
Bioinformatic prediction of active center
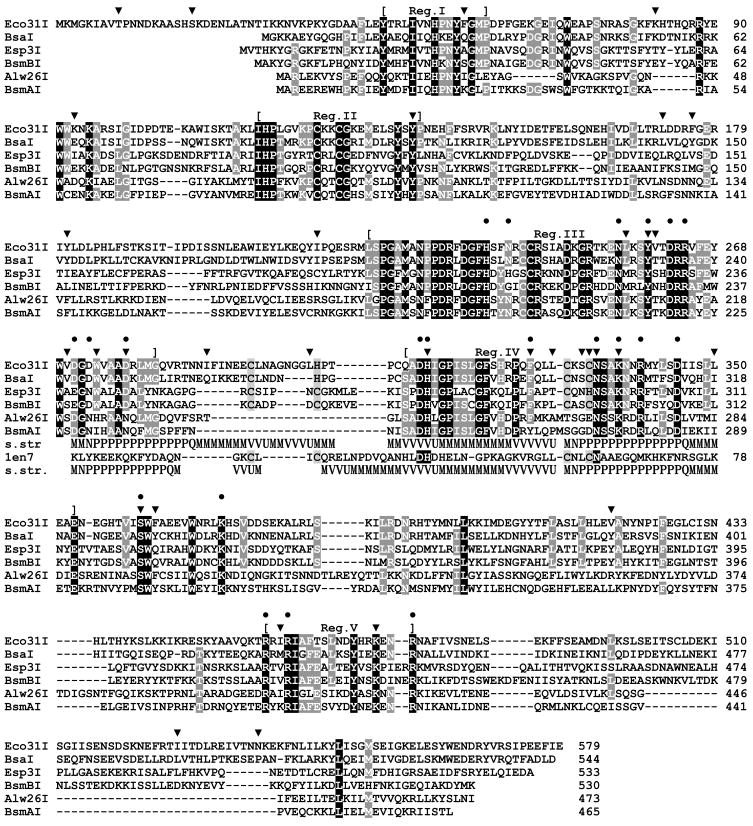
Searches of sequence databases revealed a picture typical for Type II REases: Eco31I exhibited high similarity (BLAST e-value < 7e-50) to a small group of closely related REases recognizing a common core sequence 5′-GTCTC, including Alw26I, Esp3I,17 BsaI, BsmAI, and BsmBI, also to a putative REase RxyORF2232P, and no significant similarity to other proteins. The overall level of sequence identity between the six experimentally characterized REases is low (11%), but the conserved amino acid residues are agglomerated into five conserved regions (Figure 1). Thus, these enzymes are mutually evolutionary related and their structural organization is similar. Therefore, characterization of the active site(s) of Eco31I may be extrapolated to other five enzymes.
Figure 1. Alignment of Eco31I and related REases recognizing a common pentanucleotide GTCTC.
Residues conserved in 100% and >50% sequences are indicated by white color with black and grey shading, respectively. Mutations obtained by random mutagenesis are indicated by ‘▼;’; mutations introduced by site-directed mutagenesis are indicated by ‘●’. The middle panel indicates the predicted ‘His-Me finger’ structure with the HNH-like motif, aligned to the T4 Endonuclease VII structure (1en7). Secondary structures observed in 1en7 and predicted for Eco31I are shown (helices as tubes, strands as arrows). Additionally, the common functionally important residues are shaded in T4 Endonuclease VII sequence. Residues participating in the Zn-binding site (observed in 1en7, predicted for REases) are indicated by light grey shading.
A preliminary search for the catalytic domain(s) in Eco31I was done by comparing its amino acid sequence with the sequence profiles of previously characterized protein domains using the NCBI CD-Search service.18 The search revealed a similarity of the 305-340 amino acid region to the HNH domain in Pfam and SMART databases, albeit with statistically non-significant scores (e-values 2.5 and 0.1). This region was also matched with the McrA domain (which belongs to the HNH superfamily19 in the COG and CDD databases, again with low scores (e-value 0.58 and 1.0, respectively). Another search done using InterProScan service20 at EBI indicated that the region containing residues 226-334 of Eco31I is similar to the SCOP fold of His-Me finger endonucleases, a group of enzymes with variable overall structures that nevertheless include the common HNH catalytic site (score 6.6e-4). We attempted to confirm these weak similarities using the protein fold-recognition approach. However, no methods identified any known domain in the sequence of Eco31I or in the alignment of its homologs. The only exception was the recently developed HHsearch method for HMM-HMM comparisons, which confirmed a match between the central region of Eco31I and HNH proteins (e.g. the profiles from the PFAM and CDD databases reported with P-values 3.8e-6 and 4.4e-6). However, no other domains were identified in Eco31I even with HHsearch.
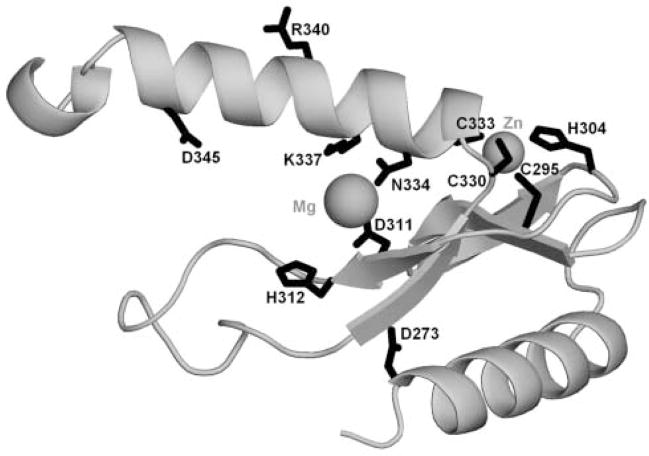
From the HMM-HMM alignment reported by HHsearch we inferred the alignment between the region 306–345 of Eco31I and the template structure of phage T4 endonuclease VII (EndoVII)21 (Figure 1) and used it to construct a putative structural model of the Eco31I active site (Figure 2).
Figure 2. Model of the ‘His-Me finger’ structure and the HNH-like active site of Eco31I (residues 271-354).
The protein backbone is shown as a grey ribbon. Side-chains of selected functionally important residues are shown in the wireframe representation and labeled. The predicted positions of Mg2+ and Zn2+ ions are indicated by balls.
The alignment and the model reveals that amino acid residues D311 and N334 in Eco31I correspond to D40 and N62 in EndoVII, respectively, and might be responsible for the cofactor (Mg2+) binding. H312 in Eco31I corresponds to H41 in EndoVII and might function as a general base, which activates a nucleophile. The central ‘N’ residue of the HNH motif, which is involved in structural stabilization, but may be absent (e.g. substituted by ‘V’ in EndoVII), is replaced by non-conserved Q325 in Eco31I. On the other hand, the structure of the predicted ‘His-Me finger’ is stabilized by a putative Zn-binding site comprising semi-conserved motifs C-X2–8-(C/H) (C295-H304 in Eco31I) and C-X2-(C/H) (C330-C333 in Eco31I), with residues that are spatially equivalent to similar Zn-binding residues C23, C26, C58 and C61 in EndoVII.21 These Cys and His residues are absent in BsmAI and Alw26I (Figure 1), indicating that these two REases probably achieve stabilization of the active site by different means. The invariant Lys residue (K337 in Eco31I) is positioned on a putative DNA-binding face of the model in such a way that it may be involved in DNA binding.
Mutagenesis of eco31IR gene
Recently, the presence of two active sites was described in the isoschizomer of Eco31I, BsaI.15 Our bioinformatic analysis of amino acid sequence of Eco31I revealed a single HNH-like active site. In order to confirm our prediction that Eco31I contains the predicted HNH active site and to clarify whether one or two active sites are present in the Eco31I, we decided to employ random mutagenesis to avoid the plausible prejudice, which amino acids are involved in the nuclease activity. We sought to identify Eco31I variants positive in DNA binding and defective in DNA restriction. We took advantage of the observation that basal expression level of Eco31I from the expression plasmid pΔ2-Eco31IR in E. coli ER2267 cells is lethal to the host in the absence of an MTase-expressing plasmid p184-Eco31IM. To select variants lacking nuclease activity, the error-prone PCR was employed to introduce mutations in eco31IR gene. The resulting DNA was used to transform E. coli ER2267 and ampicillin-resistant survivors were selected. Since some of the survived clones were expected to represent false positive mutants due to plasmid rearrangements, 384 variants were screened using PCR and 92 of them were found to contain the full-length Eco31I gene. We focused on those clones still expressing the full-length Eco31I. To identify such variants, the cells of 92 selected clones were induced, crude cell extracts were prepared and subjected to the SDS-PAGE analysis. A band corresponding to the intact Eco31I protein was prominent in 13 of 92 samples (data not shown). These 13 clones were further characterized for DNA-binding and cleavage.
Functional analysis of primary mutant enzymes
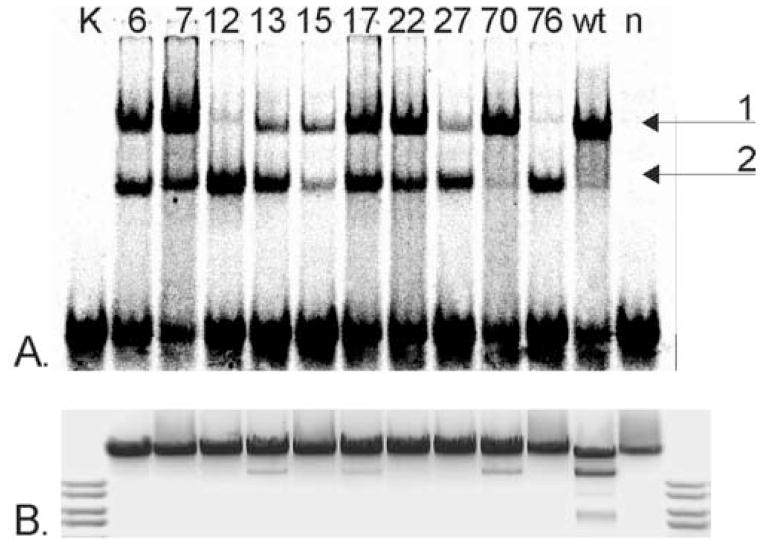
DNA-binding function of the selected mutant proteins was determined using gel-mobility shift assay. To distinguish from non-specific E. coli DNA-binding proteins in the crude extracts, the extracts obtained from mutant clones were compared with the extracts prepared from cells expressing the wild-type (wt) Eco31I and containing the expression plasmid pΔ2 without any insert gene (Figure 3A). The Eco31I non-specific DNA (negative control) was also used in the gel-mobility shift assay and no DNA binding activity was detected (data not shown). The slower migrating complexes were found in 10 cell extract samples of 13 tested (Figure 3A). All extracts carrying the mutant proteins as well as the extract carrying wt REase displayed two forms of complexes with different mobility. The slower-migrating complex 1 most likely corresponded to the full-length Eco31I bound to the target DNA. The faster migrating complex 2 corresponds to the specific complex between DNA and separately isolated DNA-binding domain, which was generated by in vitro proteolysis of the Eco31I (A. Jakubauskas, unpublished data). Presumably, the same mechanism of Eco31I specific fragmentation operates in vivo. We did not estimate the quantity of the target enzyme in the crude extract, therefore the obtained results are not directly comparable between the samples and must be evaluated only as a qualitative characteristic of the particular enzyme. Different ratios of the full-length enzyme and the proteolytically cleaved DNA-binding domain complexed with the DNA might characterize the relative sensibility of some mutant proteins to intracellular proteases, in particular that only minuscule amount of DNA-binding domain was observed in the case of wt enzyme.
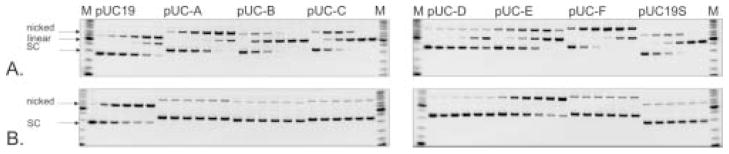
Figure 3. Functional characteristics of primary Eco31I mutants. (A.) DNA-binding, (B.) DNA restriction activity (invert image).
Mutant numbers are indicated above the lanes. K, control DNA; wt, cell extract with wt Eco31I; n, cell extract without enzyme.
Increased sensitivity to proteolysis presumably resulted from the compromised structure of some mutant enzymes. The full-length m76 mutant protein was almost incapable of binding DNA, while its DNA-binding domain exhibited significant binding. This might indicate that mutations in m76 did not affect the DNA-binding domain but had strong impact on the DNA-binding function of the full-length enzyme. The majority of the full-length m12 enzyme was found to be insoluble (data not shown). Accordingly, the band representing the full-length enzyme-DNA complex is very weak.
The relative cleavage activities of the 10 DNA-binding-positive Eco31I variants were estimated. The crude extracts were used to cleave λ DNA. Of the 10 variants tested, three (m13, m17 and m70) exhibited weak cleavage activity, while the remaining 7 variants exhibited no detectable cleavage (Figure 3B). Data on DNA binding and cleavage activity of mutants is summarized in Table 1.
Table 1.
Eco31I variants isolated by random mutagenesis.
 |
‘+’ restriction activity; ‘+/−’ partial restriction activity; ‘−’ no activity; ‘n’ nicking restriction activity; ‘FL’ full-length enzyme; ‘D’ DNA-binding domain. Amino acids conserved among all six REases are marked in bold. Amino acids located in Regions III and IV are indicated by grey shading.
DNA sequence analysis of mutant eco31IR genes
To link the different biochemical features of mutant enzymes to the specific amino acid changes, the complete nucleotide sequence of the entire eco31IR gene for 10 variants was determined. No mutants carrying a single amino acid substitution were observed – all sequenced variants carried at least two mutations (Table 1). Remarkably, every mutant contained at least one mutation in Region III (221-282 aa) and Region IV (309-353 aa) and the majority of mutations were clustered in the central part of the enzyme, where the location of the HNH-like active center was predicted. Furthermore, a set of substitutions (Y138F, Y259C, H312N, N334D, K337E, S361P and K472E) replaced amino acids that are conserved among all members of the Eco31I family (Figure 1).
Functional analysis of secondary mutant proteins
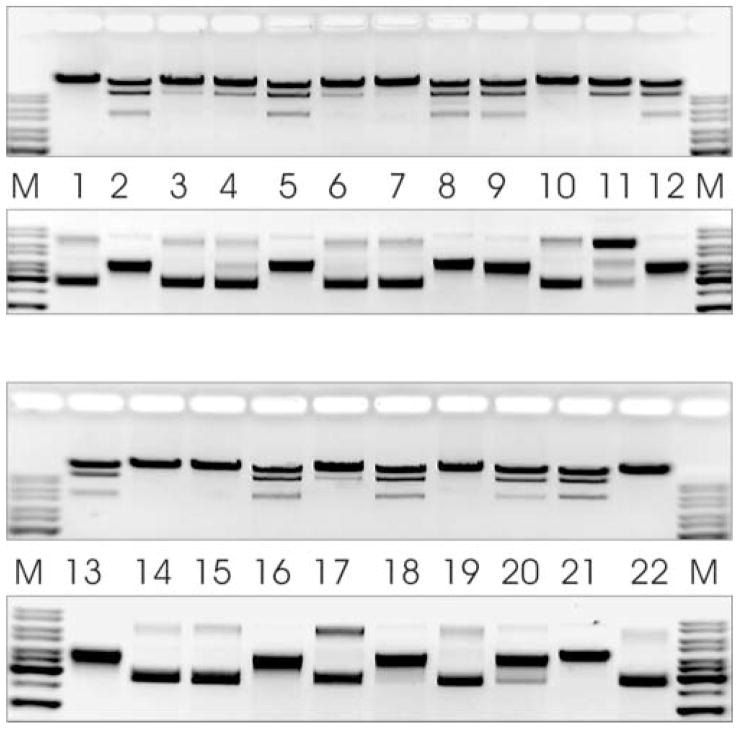
To separate individual mutations, subcloning experiments were carried out, depending on available restriction sites. Therefore, in some cases, only the elimination of some mutations from their clusters was achieved. Restriction activity of the obtained secondary mutants on λ DNA and pBR322 substrates was tested (Figure 4 and Table 1).
Figure 4. Restriction activity of secondary Eco31I mutants on λ DNA and pBR322 (invert images).
M, DNA molecular weight marker. Lane 1, m6-L303S/S332P/K337E; lane 2, m7-W274C; lane 3, m7-S361P; lane 4, m12-F326S/L350F; lane 5, m12-K472E; lane 6, m13-C333S; lane 7, m15-C333R; lane 8, m15-I459V; lane 9, m17-K82R/V270M; lane 10, m17-V270M/N334D; lane 11, m17-N334D; lane 12, m27-Y181F; lane 13, m27-Y181F/V260D; lane 14, m27-V260D/L329P; lane 15, m27-L329P; lane 16, m70-Y138F; lane 17, m70-L256P; lane 18, m76-I214T/Y259C; lane 19, m76-Y259C/D278G/I289V/F363L; lane 20, m76-D278G/I289V/F363L; lane 21, wt Eco31I; lane 22, cell extract without enzyme.
Overall analysis of restriction activities of primary and secondary mutants confirmed the predictions based on the bioinformatic analysis. Mutations that influenced the restriction activity of Eco31I were found in the Region IV (Figure 1). The N334D mutant with a substitution located in the key position of the predicted active center was found to exhibit nicking activity, prevailing to the restriction of both DNA strands. The substantial inactivation of the enzyme by C333S and C333R substitutions supported the prediction that this residue is involved in the formation of a stabilizing Zn2+-binding site. Mutations L256P and S361P are located outside the Region IV (and outside the modeled structure of the active site) but the observed reduction of restriction activity might be due to the possible distortion of the spatial arrangement of amino acids composing active center by the rigid structure of cyclic side chain of proline. The Y259C mutation (located in Region III) was found to impair the catalytic activity but only together with a set of D278G/I289V/F363L mutations.
Analysis of site-directed mutants
We did not succeed to isolate mutants harboring individual mutations Y259C, H312N and K337E located in the conserved positions among six REases by the subcloning experiments. Furthermore, the random mutagenesis was not comprehensive. Consequently, the next step towards identification of amino acids involved into the catalytic function or the formation of active center(s) was site-directed mutagenesis. The amino acids selected for alanine scanning covered all conserved positions that were found mutated after the random mutagenesis in the Region IV. Furthermore, amino acids either fully or partially conserved in the Regions III and IV among six REases recognizing common 5′-GTCTC (Figure 1) were selected. The positions H237, N240, N255, Y259, D262, D271, D273, D278, D311, H312, F326, N334, K337, R340, D345, S361 and K372 were independently changed into alanine to produce single-site mutants. Moreover, the R264D mutation was constructed to compare the resultant phenotype with the phenotype of R236D mutation in BsaI (isoschizomer of Eco31I). In addition to R236, which was claimed to be a part of the catalytic center, the R442, was proposed to participate in the second catalytic center of BsaI.15 We introduced alanine substitutions into selected conserved positions of Region V: R457, R460 and R475 (equivalent of R442 in BsaI).
Analysis of the nuclease activity on the pBR322 substrate for crude extracts from strains expressing H237A, N240A, N255A, Y259A, D262A, D271A, D278A, F326A, S361A, K372A, R457A, R460A and R475A variants revealed the full restriction activity. Hence, the corresponding residues are not involved in the restriction activity of Eco31I. Mutant F326A retained its restriction activity but was predominantly insoluble (data not shown). In the model, F326 forms a part of the ‘His-Me’ structure supporting the active site and its absence may destabilize the hydrophobic core of the catalytic domain. A similar behavior was exhibited by mutant m12 (see Table 1 and Figure 3). The remaining mutants were either inactive (D273A, H312A and N334A) or cleaved both DNA strands but accumulated nicked form of plasmid DNA (R264D, K337A and R340A) or cleaved only one DNA strand (D311A and D345A) (Figure 5). Restriction activity of the site-directed mutants confirmed the results obtained after the random mutagenesis and the additional amino acids (R264, D311, R340 and D345) influencing the catalytic function were identified after site-directed mutagenesis.
Figure 5. Restriction activity of site-directed Eco31I variants on pBR322 (invert image).
M, DNA molecular weight marker; WT, cell extract with wt Eco31I; C, cell extract without enzyme. Mutants are indicated above the lines.
Catalytic properties of the selected mutant enzymes
Lost or decreased restriction activity revealed by testing crude extracts cannot be accepted as the final enzyme characteristic and could serve only as basis for the further analysis. Therefore, we purified Eco31I variants carrying mutations R264D, D273A, D311A, H312A, N334D, K337A and D345A for detailed analysis of their nuclease activities and to determine strand specificity of the observed nicking activity.
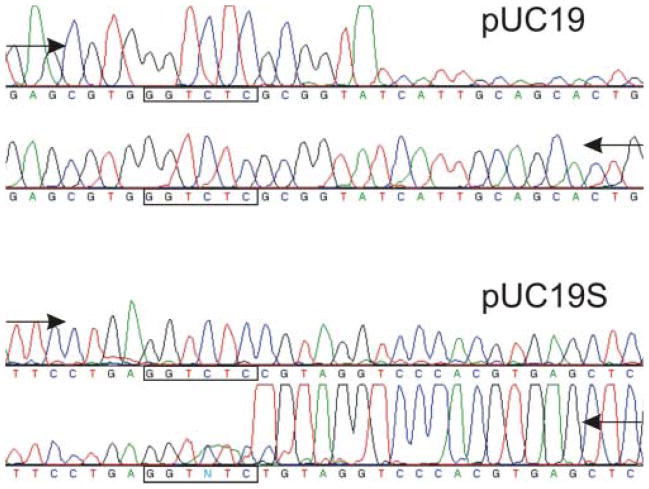
Initial run-off sequencing experiments of the pUC19 and pUC19S nicked by Eco31I-N334D revealed unexpected results. It was determined that pUC19 was nicked in the bottom strand, 5 nucleotides downstream of the cognate DNA sequence, 5′-GGTCTCNNNNN↑, and pUC19S was nicked in the top strand, 1 nucleotide downstream of the cognate DNA sequence, 5′-GGTCTCN↓ (Figure 6). These results have led us to suspect that nicking activity of Eco31I-N334D might be dependent on nucleotide content flanking cleavage positions. Therefore, additional six plasmids carrying a single Eco31I site with different flanking sequences (Table 2) were constructed. The restriction activities of all purified Eco31I variants were tested on all eight substrates. The reaction courses of Eco31I-N334D and Eco31-D311A variants are shown in Figure 7 as examples. The nicked DNA forms of each substrate (where available) were extracted from the agarose gel and analyzed by run-off sequencing. Sequencing results are presented in Table 3. The sequencing results obtained after substrate treatment with Eco31I-N334D and Eco31I-K337A were identical. Eco31I targets on pUC19 and pUC-E at first were cut on the bottom strand and in the rest of plasmids (except for pUC-D) – in the top strand. Extended incubation of the enzyme variants with the substrate resulted in the second strand being also cleaved, yielding linear DNA in all cases. pUC-D DNA was cut directly to the linear DNA form without accumulation of the nicked DNA as in the case of the wt enzyme (A. Jakubauskas, unpublished data). Sequencing results in the case of Eco31I-D345A were found to be similar to the case of Eco31I-N334D/Eco31I-K337A, with the only exception that the first cut was made in the bottom strand of pUC19S. Mutant Eco31I-D311A was found to possess very weak restriction activity compared to the mutants described above. Plasmids pUC19 and pUC-E were cut only in the bottom strand and the minuscule amount of pUC-A was cleaved in the top strand. Plasmids pUC19S, pUC-D and pUC-F were not cleaved while pUC-B and pUC-C were hardly linearized without accumulation of the nicked form. Strand specificity demonstrated by Eco31I-R264D was independent of the substrate used and the top-cleaved nicked form was accumulated prior to the bottom-strand cleavage. The plasmid pUC-F was found to be nicked at two positions. The second nick took place 12 nucleotides downstream of the cognate DNA sequence. Eco31I-D273A demonstrated weak restriction activity but cleaved all substrates to the linear DNA form without accumulation of the nicked form. Eco31I-H312A was found to be fully inactive independently of the substrate used.
Figure 6. Run-off sequencing to determine the nicking activity of Eco31I-N334D.
The drop in peak signal indicates where the DNA polymerase runs off the template at the nicked site. Arrows indicate direction of DNA synthesis. Modified DNA polymerase adds an additional adenine (A) at the end of the extension product.
Table 2.
Nucleotide sequences flanking Eco31I target.
| Plasmid | DNA sequence |
|---|---|
| pUC19 | 5′-GAGCGTGGGTCTCG↓CGGT↑ATCATT |
| pUC19S | 5′-TTCCTGAGGTCTCC↓GTAG↑GTCCCA |
| pUC-A | 5′-GCAAAAAGGTCTCC↓AAAA↑GTATTT |
| pUC-B | 5′-GCTGGCTGGTCTCT↓ACGA↑AGTCGT |
| pUC-C | 5′-GCCCAGTGGTCTCC↓AGGC↑TTCAGG |
| pUC-D | 5′-ATGCCTCGGTCTCC↓CAAG↑CGAGCT |
| pUC-E | 5′-CCGCTGTGGTCTCC↓CTTT↑AGTGAG |
| pUC-F | 5′-CTGCTTTGGTCTCT↓GATA↑GCCTTT |
Figure 7. Reaction courses of Eco31I variants on plasmid DNA substrates (invert images).
M, DNA molecular weight marker; c(enzyme) = 100 μM; c(pDNA) = 20 nM. First lane of each reaction indicates zero point. Before loading the samples were pre-heated at 75°C for 20 min to avoid shifted zones. A. Reaction course of Eco31I-N334D. Aliquots were withdrawn at 2, 5, 10, 30 and 60 min. B. Reaction course of Eco31I-D311A. Aliquots were withdrawn at 1, 2, 3, 4 and 5 h.
Table 3.
Strand specificity of Eco31I variants on different plasmid substrates.
| R264D | D311A | N334D/K337A | D345A | |
|---|---|---|---|---|
| pUC19 | 5′-GGTCTCN↓ | 5′-GGTCTCN5↑ | 5′-GGTCTCN5↑ | 5′-GGTCTCN5↑ |
| pUC19S | 5′-GGTCTCN↓ | not cleaved | 5′-GGTCTCN↓ | 5′-GGTCTCN5↑ |
| pUC-A | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ |
| pUC-B | 5′-GGTCTCN↓ | no nick | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ |
| pUC-C | 5′-GGTCTCN↓ | no nick | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ |
| pUC-D | 5′-GGTCTCN↓ | not cleaved | no nick | no nick |
| pUC-E | 5′-GGTCTCN↓ | 5′-GGTCTCN5↑ | 5′-GGTCTCN5↑ | 5′-GGTCTCN5↑ |
| pUC-F | 5′-GGTCTCN1, 12↓ | not cleaved | 5′-GGTCTCN↓ | 5′-GGTCTCN↓ |
Gel-filtration analysis of Eco31I-DNA complexes
Results obtained after random and site-specific mutagenesis indicated the only active site in Eco31I. Hence, we may predict that Eco31I is either homodimeric protein with one active site in each subunit or monomer and dimerizes in the presence of DNA.
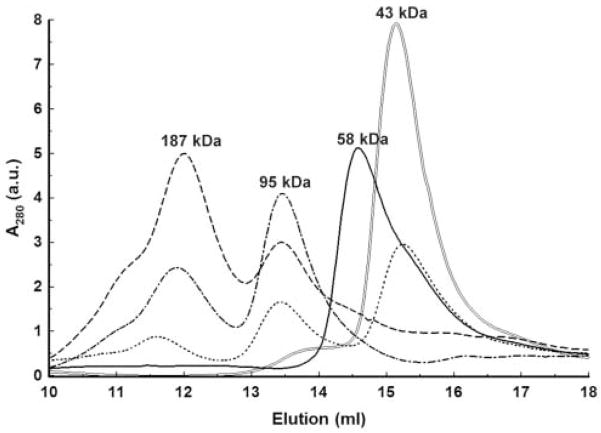
We chose gel-filtration to estimate the oligomeric state of free Eco31I in solution and the stoichiometry of Eco31I-DNA complexes. When free Eco31I was applied on column, it eluted as a single peak at volume that corresponded to an apparent molecular mass of ~58 kDa (Figure 8). The calculated molecular mass of Eco31I is 67 kDa. Therefore, this result suggests that Eco31I is a monomer in solution. Thereafter, the DNA oligoduplex was applied on column. It eluted at the volume that, relative to the calibrating proteins, yielded molecular mass of 43 kDa. The apparent molecular mass of the DNA was more than twice higher than its actual molecular mass of 15.6 kDa. This is due to the cylindrically shaped duplex having much higher frictional ratio than the spherical calibrating proteins. Gel-filtration experiments on the specific DNA and Eco31I mixtures contained constant amount of DNA (0.5 μM) and varied amounts of Eco31I from 0.25 to 2.0 μM (Figure 8). In all cases two peaks of higher molecular mass were eluted at volumes corresponding to the molecular mass of ~95 kDa and ~187 kDa. The first peak well matched to the molecular mass of one molecule of Eco31I bound to one molecule of DNA, the [1+1] complex, (58 kDa + 43 kDa = 91 kDa) and the second one corresponded to the [2+2] complex (95 kDa × 2 = 190 kDa). The DNA-Eco31I mixture with molar ratio 1:4 revealed the left-side irregularly shaped form of the peak corresponding to the [2+2] complex that indicates formation of non-specific Eco31I-DNA interaction. No DNA binding was detected using non-specific DNA (data not shown).
Figure 8. Gel-filtration of Eco31I and its complexes with DNA.
The numbers above the peaks denote the apparent molecular mass values, calculated by interpolating measured elution volumes onto the calibration curve. a.u. – arbitrary units. ‘—’ elution of free Eco31I; ‘=’ elution of free DNA; ‘■ ■ ■ ■’ elution of Eco31I-DNA (0.25–0.5 μM) complex; ‘- ■ -’ elution of Eco31I-DNA (1.0–0.5 μM) complex; ‘- - -’ elution of Eco31I-DNA (2.0–0.5 μM) complex.
DISCUSSION
Using bioinformatic methods, we predicted that Region IV, conserved in Eco31I and its homologs, forms the ‘His-Me finger’ or ‘ββα-Me finger’ structure, harboring the HNH-like active site. This structural motif was observed in endonuclease Vvn,22 homing endonuclease I-PpoI,23 Serratia nuclease,24 phage T4 endonuclease VII,21 colicins E725,26 and E9.27 The HNH active site was also predicted by bioinformatics28 and later demonstrated experimentally to be present in various Type II REases, including KpnI,29 MnlI,7 and HphI.10 Structurally, the ββα-Me finger forms an antiparallel β-hairpin followed by an α-helix, which provide a scaffold for binding of a metal ion by residues located in the first strand and in the helix. The above-mentioned enzymes have different overall three-dimensional structures, suggesting that the common catalytic module has been inserted into evolutionarily unrelated domains or independently developed different structural elaborations that stabilize the minimal ββα core.30
By using simple selection scheme we have isolated Eco31I variants obtained after random mutagenesis that displayed reduced or undetectable restriction activities and retained DNA binding abilities. Analysis of the mutations indicated the ‘hot-spot’ region, which covered two central conserved Regions III and IV of the enzyme (Figure 1). The random mutagenesis was too limited to be comprehensive and we used site-directed mutagenesis to examine the influence of the most conserved amino acids on the restriction activity. A set of substitutions, which influenced the catalytic activity of Eco31I, was identified. Eco31I variants carrying alanine substitutions of D273, H312, K337, D345 and aspartate substitutions of N334 and R264 were purified and their nuclease activity was analyzed in detail. All these residues except R264 are within our model of the ‘His-Me finger’ structure harboring the HNH motif (Figure 2)
Mutations R264D and D273A are located in the Region III (Figure 1). According to the structural prediction, semi-conserved D273 is located not in the active site, but in its vicinity, sufficiently close to enable its potential participation in DNA-binding. The invariant residue R264 is not included in our model, but it is likely that it is located not very far from the N-terminal end of the model, also at the DNA-binding side of the ‘His-Me finger’ structure. Thus, Region III could be responsible primarily for DNA binding rather than catalysis, and R264 could be involved in phosphate binding as R61 in I-PpoI.32 We compared the phenotype of Eco31I-R264D with the phenotype of the same mutation at the equivalent position in BsaI (R236D). In general, the observed restriction activity of Eco31I-R264D is comparable with the restriction activity of BsaI-R236D15. During the reaction course, the purified Eco31I-R264D generated almost exclusively the nicked DNA form, but later cleaved the second strand, thereby producing linear DNA. Only partial double-stranded DNA nuclease activity of BsaI-R236D was demonstrated, but these experiments were carried out using only cell extracts, and not a purified protein. Hence, the full characteristics of BsaI-R236D remain unknown. The D273A mutant of Eco31I exhibited strong reduction of the nuclease activity but cleaved both DNA strands as the wt enzyme, without accumulation of the nicked DNA. This result agrees with the structural prediction that D273 is not a part of the active site, but might participate in DNA binding.
Substitutions located in the Region IV, H312A and N334A were found to fully inactivate the Eco31I nuclease, in agreement with their predicted key position within the HNH catalytic motif. In the N334D mutant obtained in random mutagenesis, the DNA strands were cleaved sequentially, leading to accumulation of the nicked DNA during the reaction course. The N334D mutation introduces a charged carboxylate group instead of an uncharged amide, which may disturb the charge balance in the evolutionarily optimized metal-binding site of Eco31I. It is noteworthy that some members of the HNH superfamily, including the HphI enzyme, exhibit a ‘D’ residue at the equivalent position.10 The D311A variant of Eco31I exhibited only nicking or residual double stranded nuclease activity on some substrates. This mutation also supports the direct participation of D311 in the active site. In addition, the above-mentioned mutants exhibited no defect in DNA binding (data not shown), similarly to analogous mutations reported for other Type IIS REases from the HNH superfamily (e.g. HphI10).
Alanine mutants of Eco31I K337A, R340A and D345A cleaved DNA in the same manner as the N334D mutant. These residues are fully conserved among Eco31I homologs (Figure 1). The partial influence of these alanine mutations on the nuclease activity suggest that K337, R340 and D345 may indirectly participate in the cleavage reaction. According to our model, these residues are located in the C-terminal α-helix of the ‘ββα-Me’ structure. While D345 could be directly involved in DNA binding, the side chains of K337 and R340 are located on the opposite side of the structure and are more likely involved in stabilization of the structure than in direct interactions with the substrate.
According to our bioinformatic analysis, the predicted active center of Eco31I to be most similar to the active centers of structurally characterized EndoVII (Figure 1) and I-PpoI (data not shown). Therefore, we can suggest the catalytic mechanism of Eco31I by analogy to the mechanisms of EndoVII and I-PpoI.21,23 We propose that the cofactor (Mg2+) is coordinated by residues D311 and N334 (equivalents of D40 and N62 in EndoVII, respectively). H312 (equivalent of H41) may function as the general base, generating a hydroxyl ion for the nucleophilic attack on the scissile phosphodiester bond by activating a water molecule. K337 (equivalent of E65) together with R340 and D345 are located in close proximity to the active center and are essential for correct folding of catalytic motif (direct participation of D345 in DNA binding is also possible). We predict that Eco31I contains a Zn-binding site, which is essential for its structural integrity and that its Zn-binding site resembles the one of EndoVII, but is different from the Zn-binding sites of I-PpoI. Substitutions of C333 (to Ser and Arg), one of the four predicted Zn-binding residues in Eco31I (equivalent of C61 in EndoVII) were found to strongly reduce the Eco31I nuclease activity, in agreement with our prediction.
Kinetic studies of Type IIS REases suggested that a subgroup of these enzymes, the so called ‘short-distance cutters’, are either monomers carrying two catalytic centers or homodimers with one active center in each subunit.11 A pair of papers announced the engineering of strand-specific DNA nicking enzymes from ‘short-distance cutters’, SapI as well as BsaI, BsmAI, and BsmBI.14,15 The authors claimed the existence of two separate active sites in these enzymes. BsaI is the isoschizomer of Eco31I, so we can compare the phenotypes of mutations obtained for both enzymes. The mutant BsaI-R236D (equivalent of R264 in Eco31I) displayed top-strand nicking activity with minimal dsDNA cleavage and the other nicking variant, BsaI-R442G, displayed bottom-strand nicking activity with minimal dsDNA cleavage. These two positions were suggested as locations of two separate active centers. We did not construct an equivalent of BsaI-R442G, but performed alanine substitutions of the conserved positions R457, R460 and R475 (equivalent of R442 in BsaI). All these alanine substitutions, as well as single K472E mutation isolated after random mutagenesis did not affect the nuclease activity of Eco31I. Therefore, we cannot support the predictions made by the Xu group15 about the presence of a second active center in the corresponding location.
We propose that Type IIS REase Eco31I possesses only one catalytic domain, which exhibits the ‘ββα-Me finger’ structure and belongs to the HNH nuclease family. Based on sequence conservation, we can extend this conclusion to other closely related Type IIS REases recognizing the common core sequence GTCTC (Figure 1). In addition to mutagenesis data, complementary evidence supports the existence of only one catalytic center in Eco31I. Run-off sequencing of nicked DNA forms revealed that some mutant enzymes accumulate the nicked DNA form and introduce nicks in different DNA strands depending on the sequence flanking the cleavage positions. Such phenomenon was not described to date for any REase. It was shown that in order for scissile phosphate of DNA to reach the metal center in the HNH motif, the DNA must undergo some distortion.31 Therefore, we speculate that the order of cleavage of two strands by Eco31I is at least in part influenced by sequence-dependent DNA structure and flexibility. We found that Eco31I is a monomer in solution as judged by gel-filtration. In the presence of specific DNA, the monomer of Eco31I binds one DNA molecule forming the [1+1] complex, which dimerizes forming the [2+2] complex. HNH nucleases are known to act either as dimers: I-PpoI,32 Serratia nuclease24 and Endo VII,21 or as monomers: I-HmuI,30 or they can switch between the monomeric or dimeric forms, depending on substrate requirements, as it was found for colicin E7.25 However, the dimeric HNH nucleases with known structures produce 3′ 4 bp overhangs, while Eco31I produces 5′ 4 bp overhang, indicating that its catalytic domains may dimerize in a completely different way. For this reason, we have not attempted to construct a model of the Eco31I dimer. Nonetheless, the data presented in this work provide a solid platform for further experiments aiming at elucidation of tertiary and quaternary structure of this intriguing enzyme and the dynamics of its action.
MATERIALS AND METHODS
Bacterial strains, plasmids and reagents
The Escherichia coli strain ER2267 (New England Biolabs) was used as host for cloning experiments and expression of Eco31I (GenBank/EMBL accession no. AAM09638). E. coli cells were grown in LB broth or on LB agar at 37°C supplemented with ampicillin (0.1 mg/ml), kanamycin (0.05 mg/ml) and chloramphenicol (0.02 mg/ml) as required. An expression plasmid pUHE25-2 was kindly provided by prof. H. Bujard (ZMBH, Germany). The pΔ2, derivative of pUHE25-2 constructed by deletion of NheI-XbaI fragment, was used for the expression of Eco31I and its mutants. All enzymes, molecular biology kits, DNA and protein molecular weight markers were donated by Fermentas. [α-33P]ATP for DNA labeling was from Amersham. Oligonucleotides used for gel mobility-shift assay and PCR were synthesized by MWG-Biotech and Metabion.
DNA manipulations
Plasmid DNA preparation, DNA restriction, subcloning of DNA fragments was carried out by standard procedures18. Nucleotide sequences were generated using ‘Big-Dye’ terminator chemistry and data collected on Genetic Analyzer 3130xl (Applied Biosystems).
Construction of plasmids
A two-plasmid system was employed for the expression of R.Eco31I and its mutants. Plasmid p184-Eco31IM was constructed by cloning 3.0 kb Bsp1407I-BglII fragment, carrying both methyltransferase genes, from pEco31IRM17 into pACYC184 pre-cut with Eco32I+BamHI. Genes for Eco31I methyltransferases were expressed constitutively under control of the tetracycline resistance gene promoter. PCR was used to clone eco31IR gene from the pEco31IRM into the expression plasmid pΔ2 under control of the promoter PA1/O3/O4. Resultant plasmid was named pΔ2-Eco31IR. Amplified nucleotide sequence was verified by sequencing.
Plasmids, used as substrates for determination of strand-specificity of Eco31I variants, were constructed on the basis of pUC19S plasmid (Eco31I target in the bla gene of pUC19 was eliminated without inactivating its function and the new Eco31I target was introduced into multiple cloning site; R. Vaisvila, unpublished). pUC19S was cut with XapI+Eco72I and ligated with: 457 bp MunI-Hpy8I fragment from alw26IM gene17 resulting in pUC-A, 354 bp PdmI-EcoRI fragment from esp3IM gene17 resulting in pUC-B; 480 bp HincII-MunI fragment from sdaIR gene resulting in pUC-C; 362 bp XapI-Hpy8I fragment from sdaIR gene resulting in pUC-D; 540 bp OliI-EcoRI fragment from sdaIM gene (A. Jakubauskas, unpublished data) resulting in pUC-E and 381 bp Bst1107I-EcoRI fragment from papIM gene (A. Jakubauskas, unpublished data) resulting in pUC-F.
PCR mutagenesis
Random mutagenesis
We used an error-prone random mutagenesis technique that overlies the bias in transition: transversion ratio.35 This method consists of two compensatory PCR steps. Plasmid pΔ2-Eco31IR served as a matrix in the first PCR and 0.1 mM MnCl2 was added to the standard reaction buffer. The resultant PCR fragment was used as matrix in the second PCR where 0.1 mM dITP was added to the reaction buffer. Primers 5′-TCGTCTTCACCTCGAGAAAATTTATC and 5′-ATCTATCAACAGGAGTCCA, flanking the whole eco31IR gene, were used in the both reactions. The final PCR fragment was cut with XhoI+BglII and cloned back to the pΔ2-Eco31IR pre-cut with the same enzymes.
Site directed mutagenesis
Megaprimer method36 was employed to introduce the selected mutations into eco31IR gene. The resultant PCR products were cloned and their nucleotide sequences were verified by sequencing. Thereafter, DNA fragments carrying required mutation were subcloned back to the pΔ2-Eco31IR.
Transformation and preparation of cell extracts
Library of restriction-deficient eco31IR genes was obtained by electro-transformation of ER2267 strain. Electro-transformation was carried out using 2.5 kV in ‘E.coli Pulser’ (Bio-Rad). Electro-competent and chemically competent cells were prepared using standard procedures.34 Transformants were plated onto LB agar plates with appropriate antibiotics for the plasmid selection. Individual transformants after selection were inoculated into 5 ml LB broth with appropriate antibiotics. Cells were grown until late log phase and then induced 3 h with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). The cell extracts were prepared as described37 and used for gel-mobility shift assay and determination of restriction activity in vitro.
Gel-mobility shift assay
Two complementary single stranded oligonucleotides were annealed to give Eco31I-specific DNA 5′-AGCTTCGTGGGTCTCGCGGTATCAGATC with 20 bp stem and 4 nucleotide protruding 5′-ends. Specific DNA sequence (underlined) was substituted by GGCCAC to give a non-specific DNA. 5′-termini of probes were labeled with T4 polynucleotide kinase by [γ-33P] dATP. The labeled DNA (final concentration 1 nM) was 20 min incubated with different amounts of the crude extracts in TAE buffer (40 mM Tris- acetate (pH 8.0), 1 mM EDTA) supplemented with 0.1 mg/ml BSA in total 20 μl reaction volumes at 25°C. Afterwards, reactions were loaded onto 8 % polyacrylamide non-denaturing gel (29:1 acrylamide/bis-acrylamide) and fractionated in TAE buffer. Results were visualized by Cyclone™ Storage Phosphor System and analyzed by using OptiQuant™ Image Analysis Software (Packard).
Purification of proteins
E. coli 2267 [pΔ2-Eco31IR+p184-Eco31IM] cells were grown with aeration in LB broth. Expression of the gene coding for R.Eco31I and its mutants was induced by adding of IPTG to the final 1 mM concentration to the culture at OD600 of ~0.5. After 3 h induction the cells were harvested by centrifugation and stored at −20°C. All further steps were carried out at 4°C. Biomasses were thawed in 10 mM K-phosphate (pH 7.0), 1 mM EDTA, 7 mM 2-mercaptoethanol, 100 mM KCl buffer. Then cells were disrupted by sonication, cell debris was removed by centrifugation and obtained supernatants were sequentially loaded on Heparin-Sepharose, SP-Sepharose, Blue-Sepharose, CM-Sepharose (Amersham) and Hydroxyapatite (Calbiochem) columns. Eluted Eco31I fractions assayed for activity and by SDS-PAGE, were pooled and dialyzed against storage buffer (20 mM Tris-HCl (pH 7.5), 200 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol) and stored at −20°C.
The obtained proteins were > 95% homogeneous as judged by SDS-PAGE analysis. Protein concentrations were determined spectrophotometrically by absorbance at 280 nm, using calculated extinction coefficient 87540 M−1cm−1. The concentrations are given in the terms of monomeric form of protein.
Restriction activity in vitro and determination of strand specificity
Restriction activity in vitro was assayed by incubating pBR322 with 1 μl cell extract in Tango™ buffer (Fermentas) in total 30 μl volume for 30 min at 37°C and visualizing by agarose gel electrophoresis. The conditions for testing restriction activity of purified Eco31I mutant proteins and determination of their strand specificity were as follows: 20 nM of plasmid DNA and a particular amount of enzyme were incubated in G+ buffer (Fermentas) in total 70 μl volumes at 37°C. The probe without added protein served as zero point. At various time points the aliquots were stopped by adding the loading dye solution and heating at 75°C for 10 min. The reaction products were analyzed by agarose gel electrophoresis. The nicked DNA form was then gel-purified using ‘DNA Extraction kit’ and used as a template for runoff sequencing.
Gel-filtration
Gel-filtration of the purified Eco31I and its mixtures with specific and non specific DNA (see Gel-mobility shift assay) was performed at room temperature on the AKTA FPLC system using Superdex 200 HR column (Amersham-Pharmacia) pre-equilibrated with 40 mM Tris-acetate (pH 8.0), 150 mM Na-acetate, 2 mM EDTA. The samples were prepared in 200 μl of the same buffer. Elution from the column was monitored by measuring absorbance at 280 nm. The calibration curve was established using Gel-filtration Calibration kits (Amersham-Pharmacia). The molecular masses of Eco31I, DNA and their complexes were calculated by interpolating their elution volumes onto the calibration curve.
Bioinformatic analysis
Sequence searches to identify homologs of Eco31I were carried out by PSI-BLAST38 and their alignment was done via the interactive Web server NPS@39. Identification of regions in the Eco31I sequence that exhibit sequence similarity to known domains was carried out by searching the Pfam40, SMART41, COG42, CDD43 and SUPERFAMILY44 databases. Initially, the original tools implemented in each of these databases were used to carry out sequence-vs-profile comparison, followed by searches with the HHsearch method45. HHsearch allows for very sensitive sequence searches by calculating Hidden Markov Models both for the query sequence and for the families in the database, and by performing HMM-vs-HMM comparison. Additionally, protein fold-recognition analysis and prediction of secondary structure and protein disorder was carried out via the GeneSilico meta-server46.
Modeling of the active site region of Eco31I was carried out based on the alignment reported by the HHsearch method, using the structure of phage T4 endonuclease VII (1en7 in Protein Data Bank) as the template. The ‘FRankenstein’s monster’ method was used to optimize the sequence-structure compatibility47.
Acknowledgments
Authors thank Fermentas for continuous support. This work was partly supported by Lithuanian State Science and Studies Foundation grant No 208 (to A.J.). J.M.B. was supported by the NIH (Fogarty International Center grant R03 TW007163-01). Authors thank Dr. Egle Cesnaviciene and Dr. Daumantas Matulis for linguistic help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci U S A. 1992;89:4275–9. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 4.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10570–5. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaremba M, Urbanke C, Halford SE, Siksnys V. Generation of the BfiI restriction endonuclease from the fusion of a DNA recognition domain to a non-specific nuclease from the phospholipase D superfamily. J Mol Biol. 2004;336:81–92. doi: 10.1016/j.jmb.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J Mol Biol. 2003;326:1051–64. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 7.Kriukiene E, Lubiene J, Lagunavicius A, Lubys A. MnlI - The member of H-N-H subtype of Type IIS restriction endonucleases. Biochim Biophys Acta. 2005;1751:194–204. doi: 10.1016/j.bbapap.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kriukiene E. Domain organization and metal ion requirement of the Type IIS restriction endonuclease MnlI. FEBS Lett. 2006;580:6115–22. doi: 10.1016/j.febslet.2006.09.075. [DOI] [PubMed] [Google Scholar]
- 9.Azarinskas A, Maneliene Z, Jakubauskas A. Hin4II, a new prototype restriction endonuclease from Haemophilus influenzae RFL4: discovery, cloning and expression in Escherichia coli. J Biotechnol. 2006;123:288–96. doi: 10.1016/j.jbiotec.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Cymerman IA, Obarska A, Skowronek KJ, Lubys A, Bujnicki JM. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006;65:867–76. doi: 10.1002/prot.21156. [DOI] [PubMed] [Google Scholar]
- 11.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–35. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J Biol Chem. 2002;277:4024–33. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 13.Armalyte E, Bujnicki JM, Giedriene J, Gasiunas G, Kosinski J, Lubys A. Mva1269I: a monomeric type IIS restriction endonuclease from Micrococcus varians with two EcoRI- and FokI-like catalytic domains. J Biol Chem. 2005;280:41584–94. doi: 10.1074/jbc.M506775200. [DOI] [PubMed] [Google Scholar]
- 14.Samuelson JC, Zhu Z, Xu SY. The isolation of strand-specific nicking endonucleases from a randomized SapI expression library. Nucleic Acids Res. 2004;32:3661–71. doi: 10.1093/nar/gkh674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Z, Samuelson JC, Zhou J, Dore A, Xu SY. Engineering strand-specific DNA nicking enzymes from the type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J Mol Biol. 2004;337:573–83. doi: 10.1016/j.jmb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Butkus V, Bitinaite J, Kersulyte D, Janulaitis A. A new restriction endonuclease Eco31I recognizing a non-palindromic sequence. Biochim Biophys Acta. 1985;826:208–12. doi: 10.1016/0167-4781(85)90008-9. [DOI] [PubMed] [Google Scholar]
- 17.Bitinaite J, Mitkaite G, Dauksaite V, Jakubauskas A, Timinskas A, Vaisvila R, Lubys A, Janulaitis A. Evolutionary relationship of Alw26I, Eco31I and Esp3I, restriction endonucleases that recognise overlapping sequences. Mol Genet Genomics. 2002;267:664–72. doi: 10.1007/s00438-002-0701-6. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bujnicki JM, Radlinska M, Rychlewski L. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to betabetaalphaMe endonucleases. Mol Microbiol. 2000;37:1280–1. doi: 10.1046/j.1365-2958.2000.02010.x. [DOI] [PubMed] [Google Scholar]
- 20.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, Copley R, Courcelle E, Das U, Durbin R, Fleischmann W, Gough J, Haft D, Harte N, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McDowall J, Mitchell A, Nikolskaya AN, Orchard S, Pagni M, Ponting CP, Quevillon E, Selengut J, Sigrist CJ, Silventoinen V, Studholme DJ, Vaughan R, Wu CH. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–5. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raaijmakers H, Vix O, Toro I, Golz S, Kemper B, Suck D. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 1999;18:1447–58. doi: 10.1093/emboj/18.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–25. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ, Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat Struct Biol. 1999;6:1096–9. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]
- 24.Miller MD, Tanner J, Alpaugh M, Benedik MJ, Krause KL. 2.1 A structure of Serratia endonuclease suggests a mechanism for binding to double-stranded DNA. Nat Struct Biol. 1994;1:461–8. doi: 10.1038/nsb0794-461. [DOI] [PubMed] [Google Scholar]
- 25.Cheng YS, Hsia KC, Doudeva LG, Chak KF, Yuan HS. The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J Mol Biol. 2002;324:227–36. doi: 10.1016/s0022-2836(02)01092-6. [DOI] [PubMed] [Google Scholar]
- 26.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 27.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999;6:243–52. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 28.Bujnicki JM, Radlinska M, Rychlewski L. Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem Sci. 2001;26:9–11. doi: 10.1016/s0968-0004(00)01690-x. [DOI] [PubMed] [Google Scholar]
- 29.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–35. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 31.Mate MJ, Kleanthous C. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J Biol Chem. 2004;279:34763–9. doi: 10.1074/jbc.M403719200. [DOI] [PubMed] [Google Scholar]
- 32.Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 33.Pottmeyer S, Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992;223:607–15. doi: 10.1016/0022-2836(92)90977-r. [DOI] [PubMed] [Google Scholar]
- 34.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 4. John Wiley & Sons, Inc; 1987. [Google Scholar]
- 35.Fenton C, Xu H, Petersen EI, Petersen SB, el-Gewely MR. Random mutagenesis for protein breeding. Methods Mol Biol. 2002;182:231–41. doi: 10.1385/1-59259-194-9:231. [DOI] [PubMed] [Google Scholar]
- 36.Brons-Poulsen J, Nohr J, Larsen LK. Megaprimer method for polymerase chain reaction-mediated generation of specific mutations in DNA. Methods Mol Biol. 2002;182:71–6. doi: 10.1385/1-59259-194-9:071. [DOI] [PubMed] [Google Scholar]
- 37.Whitehead PR, Brown NL. A simple and rapid method for screening bacteria for type II restriction endonucleases: enzymes in Aphanothece halophytica. Arch Microbiol. 1985;141:70–4. doi: 10.1007/BF00446742. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–50. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 40.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–6. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–9. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 46.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–7. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosinski J, Cymerman IA, Feder M, Kurowski MA, Sasin JM, Bujnicki JM. A “FRankenstein’s monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins. 2003;53(Suppl 6):369–79. doi: 10.1002/prot.10545. [DOI] [PubMed] [Google Scholar]