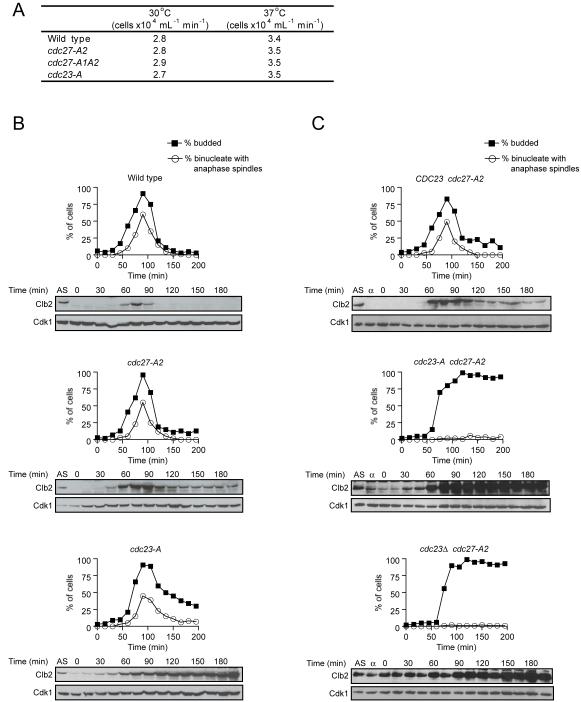
Figure 4. Cdc27 and Cdc23 mutants have mild individual defects in vivo and together are synthetically lethal.
(A) Log-phase growth rates, at 30°C and 37°C, of strains in which the indicated mutant gene replaced the endogenous gene. Results are representative of three individual experiments.
(B) Asynchronous cultures of the indicated cells (AS timepoint) were treated with α factor (1 μg/ml) for 2.5 h. α factor was then washed out (zero time point) and cells were harvested at the indicated times. α factor was added back after 90% of the cells were budded. Parallel samples were analyzed directly for budding index (black squares) and fixed, treated with zymolyase, and stained with DAPI to measure chromosome segregation, and with anti-tubulin antibodies to measure spindle elongation. We counted binucleate cells that had elongated spindles (open circles). Additional samples were prepared for western blotting with anti-Clb2 antibodies and anti-Cdk1 (as a loading control).
(C) Asynchronous cultures of the indicated PGAL1-CDC23 strains in galactose-containing media (AS timepoint) were treated with α factor for 3 h (α time point), after which dextrose was added for 2 h to turn off expression of wild-type CDC23. α factor was washed out (zero time point) and cells released into dextrose-containing media. Samples were taken at the indicated times for analysis of budding index, chromosome segregation, spindle formation, and Clb2 levels as in (B).