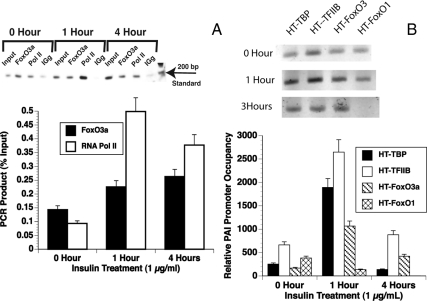
Figure 5.
FoxO3a is present on the PAI-1 promoter in BAE and T47D cells. A, BAE cells were treated with 1 μg/ml insulin for 0, 1, or 4 h. They were then treated with formaldehyde, and the nuclei were prepared and sonicated to produce fragments between 100 and 800 bp as described (Materials and Methods). The sonicated chromatin was precleared with BSA and herring sperm-DNA blocked protein A sepharose and incubated with antibody to FoxO3a (Upstate Biotechnology) or RNA polymerase II (Santa Cruz Biotechnology). The complexes were collected using BSA- and herring sperm-DNA-blocked protein A sepharose. After extensive washing, the samples were de-cross-linked and phenol-chloroform extracted and precipitated with ethanol. PCR with primers to the PAI-1 proximal promoter was performed. Top, SYBR green fluorescence was visualized using the Typhoon Trio laser scanner. Bottom, Quantitation of three experiments using ImageQuant software. B, T47D cells were transfected with plasmids expressing HT-TBP, TFIIB, FoxO1, or FoxO3a fusion proteins. The cells were treated with1 μg/ml insulin for 0, 1, or 4 h. They were then treated with formaldehyde, and the nuclei were prepared and sonicated to produce fragments between 100 and 800 bp as described (Materials and Methods). The sonicated chromatin was incubated with HaloLink resin and the complexes were collected by centrifugation and washed extensively. The samples were de-cross-linked and phenol-chloroform extracted and precipitated with ethanol. PCR with primers to the PAI-1 proximal promoter was performed. Top, SYBR green fluorescence of semiquantitative PCR was visualized using the TyphoonTrio laser scanner. Bottom, Analysis of three experiments using qPCR. The amount of DNA in each sample was determined in comparison with a standard curve. They were then compared with the value for the lowest point (HT-FoxO1 at 4 h) that was arbitrarily set equal to 1. Pol II, Polymerase II.