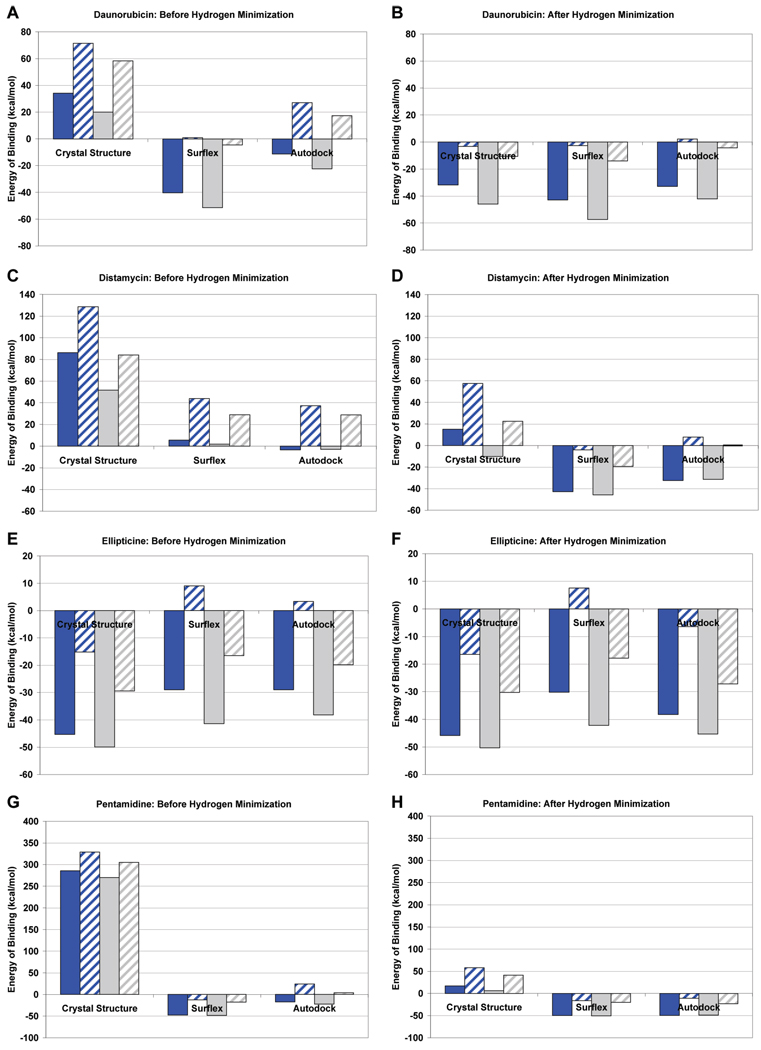
Figure 7.
Calculated energy of binding by Macromodel for the crystal pose, Autodock top ranked pose, and Surflex top ranked pose for various ligands using the Amber* and OPLS2005 force fields with and without implicit water solvation: solid blue = OPLS2005, no implicit water solvation; blue with hatches = OPLS2005, with implicit water solvation; solid gray = Amber*, no implicit water solvation; gray with hatches = Amber*, with implicit water solvation. (A) and (B) daunorubicin, before and after hydrogen minimization, respectively; (C) and (D) distamycin, before and after hydrogen minimization, respectively; (E) and (F) ellipticine, before and after hydrogen minimization, respectively; and (G) and (H) pentamidine, before and after hydrogen minimization, respectively.