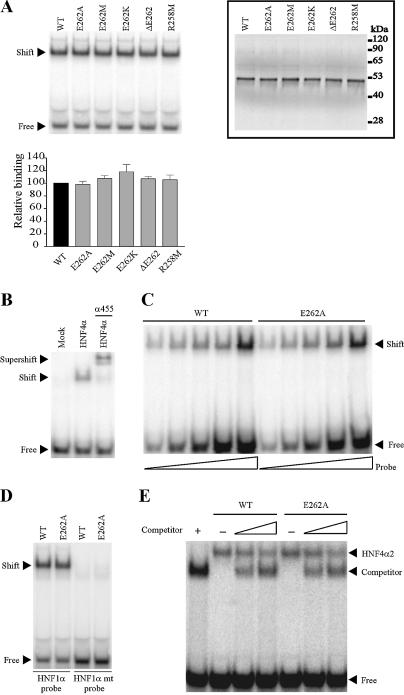
Figure 3.
Mutations of E262 and R258 residues do not impair HNF4α2 DNA binding. (A) DNA binding of HNF4α2 mutants to the 32P-labelled HNF4α response element of the HNF1α promoter (HNF1 site). Control of in vitro synthesis of wild-type and mutated HNF4α2, used in EMSA, is shown in the insert (values on the right end indicate molecular size markers). The graph indicates means ± SE of mutated HNF4α2 DNA binding relative to that of the wild-type protein from three independent experiments. (B) Specificity of binding. Unprogrammed reticulocyte lysate (mock) yielded no shifted band. Supershifting was performed in the presence of the specific α455 HNF4α antiserum. (C) EMSA performed with a constant amount of HNF4α2 WT or HNF4α2-E262A and increasing amounts of labelled HNF1 probe. (D) HNF4α2-E262A did not bind as a monomer to the half-site of the HNF4α response element (HNF1 mt). (E) Competition experiments with COUP-TFII ΔAB. EMSA were performed on the HNF4 response element of the apoCIII promoter (CIIIB site) using in vitro synthesised HNF4α2 WT or HNF4α2-E262A and increasing amounts of the competitor COUP-TFII ΔAB. The amount of reticulocyte lysate in each lane was held constant by the appropriate addition of unprogrammed lysate. The positions of HNF4α2 and COUP-TFII ΔAB homodimers bound to DNA are indicated.