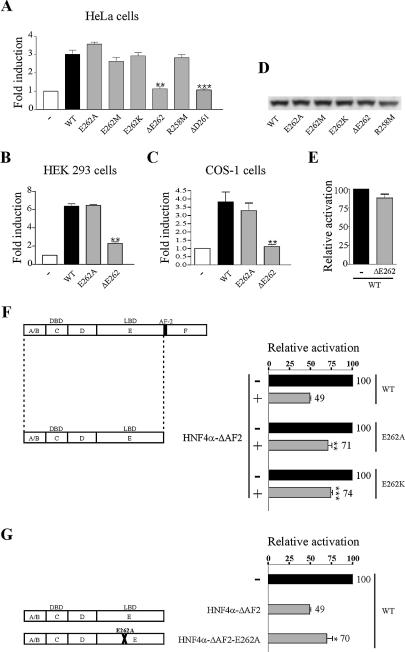
Figure 5.
Deletion of E262 strongly affects HNF4α2 transcriptional activity. HeLa (A), HEK 293 (B) and COS-1 cells (C) were transiently transfected with 12.5 ng of expression vector for wild-type or mutated HNF4α2 or the corresponding empty vector (–) together with 250 ng of HNF1α promoter construct. Fold induction refers to the activity with no HNF4α2 derivative (–), which was set to 1. Results are means ± SE of three independent experiments performed in triplicate. **, P = 0.0015, 0.0060 and 0.0018 for the ΔE262 mutant in (A–C), respectively; ***, P < 0.0001 for the ΔD261 mutant in (A). (D) Western blotting of HeLa cell extracts. (E) HNF4α-ΔE262 does not exhibit a dominant-negative activity on wild-type HNF4α. COS-1 cells were transfected as in (C), except that equal amounts of wild-type HNF4α and HNF4α-ΔE262 or control vector (–) were co-transfected. (F and G) Effects of substitution mutations on the dominant-negative activity of HNF4α-ΔAF-2. COS-1 cells were transfected as in (C), except that in (F) plasmids expressing wild-type, E262A or E262K HNF4α were co-transfected with an equal amount of vector expressing HNF4α-ΔAF-2 or the control vector (–), whereas in (G) pcDNA3 HNF4α2 WT was co-transfected with an equal amount of vectors expressing HNF4α-ΔAF-2 or HNF4α-ΔAF-2-E262A or the control vector (–). Activation of the HNF1α promoter is expressed relative to that obtained when only full-length proteins were expressed. Results are means ± SE of three independent experiments performed in triplicate. **, P = 0.0040 in (F); ***, P < 0.0001 in (F); *, P = 0.0278 in (G).