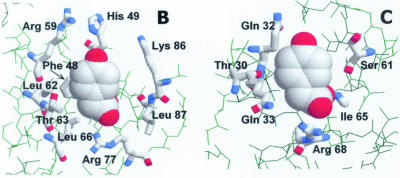
Figure 10.
Model of ligand-bound HpaR structure. (A) Amino acid sequence alignment of the HpaR and MarR proteins. Residues corresponding to the SAL-A type binding site are shown in bold. Residues corresponding to the SAL-B type binding site are represented in lower case. The putative HpaR DNA recognition helix is boxed. Alignment was done with the BESTFIT program. (B) Detail of the ‘SAL-A’ type 4HPA binding site. The ligand is depicted in spacefill representation and interacting residues are shown in thick wireframe format. Note that 4HPA could also bind in an alternative orientation, rotated 180° around the z-axis. (C) Detail of the ‘SAL-B’ type 4HPA binding site. The scheme representation is as indicated for (B).