Abstract
Affinity capillary electrophoresis (ACE) was used to study interactions of small, highly sulfated, aromatic molecules with antithrombin (AT). The high charge density of the small molecules induces differential migration of the complex resulting in a versatile method of assessing binding affinities, nature of interactions and site of binding on the inhibitor. Scatchard analysis of the interaction of three tetrahydroisoquinoline-based multiply sulfated molecules with AT results in monophasic profiles with affinities in the range of 40 – 60 μM in 20 mM sodium phosphate buffer, pH 7.4. For a pentasulfated molecule, a biphasic profile with affinities of 4.7 μM and 30 μM was observed. Measurement of KD as a function of ionic strength of the medium indicated that ionic and non-ionic forces contribute 2.4 and 1.9 kcal/mol, respectively, at pH 7.4 and 100 mM NaCl. Competitive binding studies show that the tetrahydroisoquinoline-based molecules do not compete with a high-affinity heparin pentasaccharide. In contrast, the affinity of these tetrahydroisoquinoline derivatives decreases dramatically in the presence of an extended-heparin binding site ligand. Overall, ACE analysis of small, sulfated aromatic molecules interacting with AT is relatively easy and obviates the need for an external signal, e.g., fluorescence, for monitoring the interaction. In addition to affording biochemical knowledge, the small sample requirement and fast analysis time of ACE could be particularly advantageous for high throughput screening of potential anticoagulants.
Keywords: affinity capillary electrophoresis, antithrombin, capillary electrophoresis, interaction, synthetic sulfated molecules
1 Introduction
Antithrombin (AT) is a plasma glycoprotein and a major inhibitor of thrombin, factor Xa, and factor IXa, three enzymes that play pivotal roles in the blood clotting. Yet, AT is a rather poor inhibitor of all the three enzymes under physiologic conditions.1–3 Heparin, a carbohydrate polymer prepared from animals, especially from pig and bovine, induces AT to become a potent inhibitor of the three enzymes, which is the reason for heparin’s clinical use as an anticoagulant. Clinically used heparin, appropriately named unfractionated heparin, is highly polyanionic preparation consisting millions of polysaccharide chains, each with a differing composition and structure. This structural heterogeneity is known to be associated with multiple adverse effects including hemorrhage, thrombocytopenia, osteoporosis and variable patient response,4 which introduces challenges in their usage, especially in elderly population. In addition, the animal origin of heparin introduces another set of challenges, as demonstrated by the recently observed contamination with oversulfated chondroitin sulfate.5,6
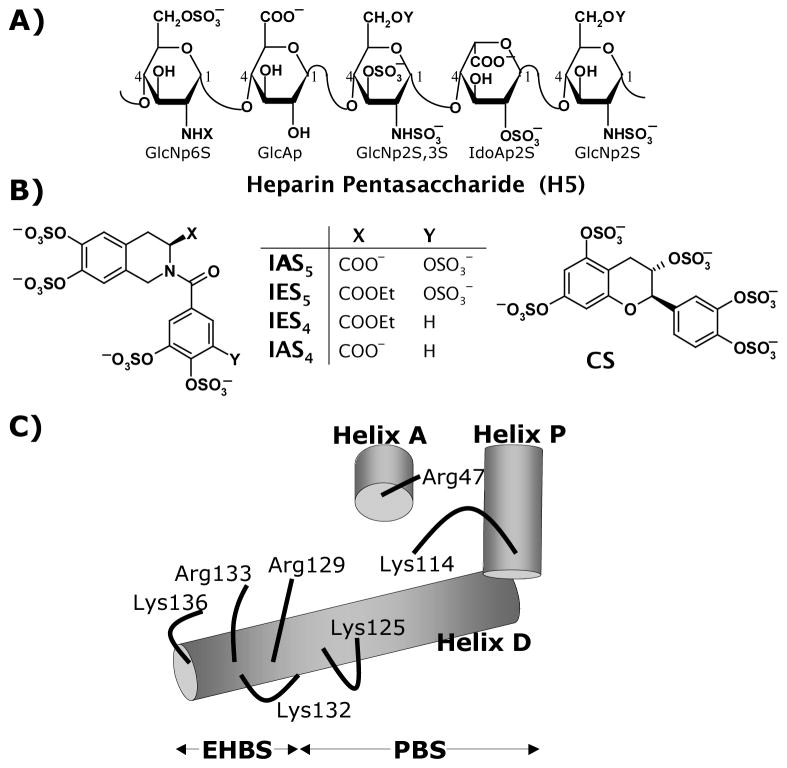
Heparin enhances the potency of AT by inducing a major conformational change in the glycoprotein, which increases the inhibitor’s activity nearly 600-fold.7,8 Despite the massive structural heterogeneity of unfractionated heparin, only selected sequences, e.g., those containing →4) β-D-GlcNp6S (1→4) β-D-GlcAp (1→4) β-D-GlcNp2S,3S (1→ (Fig. 1A), can bring about this activation suggesting phenomenonal specificity of action. Polysaccharide chains containing this sequence bind to an electropositive region in AT called the heparin-binding site (HBS), which is now understood to consist of a pentasaccharide binding site (PBS) and an extended heparin binding site (EHBS) (Fig. 1C).9,10 Interaction in the PBS appears to be critical for the induction of 600-fold activation in AT.
Figure 1.
A) Structure. of heparin pentasaccharide H5; B) Structure of non-saccharide activators of antithrombin, IAS5, IES5, IES4, and IAS4 and (+)-CS. C) Model of heparin binding site in antithrombin. Numbers in H5 structure refer to positions of carbon atoms, whereas X and Y can be either –COCH3 or –SO3− groups. The heparin-binding site in C) can be thought of as a combination of the pentasaccharide-binding site (PBS) and extended heparin-binding site (EHBS). The PBS is formed by Arg47, Lys114, Lys125 and Arg129 residues of helices D, A and P, while the EHBS is formed by Arg129, Lys132, Arg133 and Lys136 residues of helix D. See text for details.
To devise an alternative anticoagulation approach based on AT so as to avoid the problems associated with heparin therapy, we initiated a synthetic medicinal chemistry program some time ago and designed several small, non-sugar, highly sulfated, aromatic molecules.9,11–13 These small, synthetic molecules are more accessible than heparins and are likely to offer special advantages such as greater non-ionic binding energy9 in AT recognition and/or greater hydrophobicity for possible oral delivery. An interesting feature of the highly sulfated, aromatic molecules is that they simultaneously possess both hydrophobic and anionic character. Recently, we synthesized four tetrahydroisoquinoline-based potential AT activators as mimics of a heparin trisaccharide.14 Of the four, only IAS5 (Fig. 1B) was found to activate antithrombin nearly 30-fold,15 which was significantly higher than our first generation designs.9,11–13
IAS5 represents one of the few small, synthetic, non-sugar molecules that bind and activate AT, and may pave the way to more potent activators for use as anticoagulant drugs. To understand the nature of interaction of these interesting molecules, we needed a powerful biophysical technique. It is difficult to measure the AT binding affinities of such small molecules because the highly sulfated molecules typically do not induce significant differential change in AT absorption or fluorescence, nor do they significantly alter the inhibitor’s magnetic resonance properties. Other methods used in the study of heparin – antithrombin interactions, such as isothermal titration calorimetry,16 affinity chromatography,17,18 and surface plasmon resonance19,20 are not as useful for detailed small molecular interaction studies. In contrast, capillary electrophoresis (CE) offers high speed, excellent resolution, low sample size and good flexibility. Additionally, affinity capillary electrophoresis (ACE) has been used to study AT-polysaccharide interactions, especially with heparin and fucoidan.21–25 In this paper, we describe the application of ACE to understand the nature of small highly sulfated molecules binding to AT at a molecular level. Our work suggests that ACE is highly effective in measuring AT equilibrium dissociation constants under physiological conditions and is able to decipher the site of binding as well as the ionic/non-ionic nature of interaction. Additionally, we have discovered an interesting synergistic mechanism of these small molecules binding to AT in the presence of heparin pentasaccharide (Fig. 1).
2 Materials and Methods
2.1 Chemicals and Reagents
Human plasma AT (MW 58,000) was purchased from Haematologic Technologies, Inc (Essex Junction, VT) and used as such. IAS4, IAS5, IES4, IES5 and catechin sulfate ((+)-CS) (Fig. 1B) were synthesized and characterized in our laboratory.9,14,15 Heparin pentasaccharide H5 (Fig. 1A) was a gift from Professor Steven Olson (University of Illinois – Chicago). High purity water (18.2 megohms), purchased from Fisher Scientific (Agawam, MA), was used to prepare all buffers. 20 mM sodium phosphate buffer, pH 7.4, possessing ionic strength of 0.035 was used as run electrolyte. Buffers with higher ionic strengths (I = 0.045, 0.055 and 0.085) were prepared by adding 10, 20 or 50 mM NaCl to the 20 mM sodium phosphate buffer and adjusting the pH to 7.4.
2.2 Experimental Procedure
The interaction of AT with sulfated molecules was studied using a P/ACE MDQ™ Beckman capillary electrophoresis system (Fullerton, CA). Electrophoresis was performed at a constant voltage of 8 or 10 kV using an uncoated fused silica capillary (I.D. 50 or 75 μm) of 31.2 or 40.2 cm total length with a 5 mm detection window at 21.0 or 30.0 cm length, respectively from the injection point. During electrophoresis, the capillary was held at a constant temperature of 25 °C. A sequential wash of 1 M HCl for 10 min, high purity water for 3 min, 1 M NaOH for 10 min, and high purity water for 3 min at 20 psi was used to activate a capillary. Before individual electrophoretic runs, the capillary was rinsed with the run buffer for 3 min at 20 psi.
A 43 μM AT stock solution was prepared in 20 mM sodium phosphate buffer, pH 7.4, containing 100 mM NaCl, 0.1 mM EDTA, and 0.1% PEG 8000. Stock solutions of ≥11 mM IAS4, IAS5, IES4 and IES5 (Fig. 1B) were prepared in high purity water. Run electrolytes containing these molecules at desired concentrations were made through a ≥10-fold dilution with 20 mM sodium phosphate buffer, pH 7.4. Naphthol was used as neutral marker. Samples containing 0.86 μM AT and naphthol (300-fold diluted from its stock in water) were injected at the anodic end using 0.5 psi pressure for 4 s and detected at the cathodic end at a wavelength of 214 nm.
2.3 Quantitative model of the ACE method
The basic principle and analysis of ACE has been elegantly described by Whitesides and co-workers.26 Briefly, if and are the AT’s electrophoretic mobility in a reference buffer and in a buffer containing our ligand, respectively, then the expression represents the change in the mobility of AT due to the presence of the ligand. Under rapid equilibration condition, equation 1 can be derived as the electrophoretic version of the traditional Scatchard equation.27
| (1) |
In this equation KA is apparent equilibrium association constant, is the maximal change in mobility of AT when saturated with ligand L ([L] ≫ [AT]). Thus, a plot of Δμep/[L] versus Δμep will provide a value of KA from its slope. The electrophoretic mobility of AT was calculated from equation 2.26 In this equation, Le is the effective separation length of the capillary, Lt is the total length of the capillary, tAT is the migration time of AT, tnm is the migration time of the neutral marker, and V is the applied voltage during the separation process.
| (2) |
3 Results
3.1 AT Interaction Study Using ACE
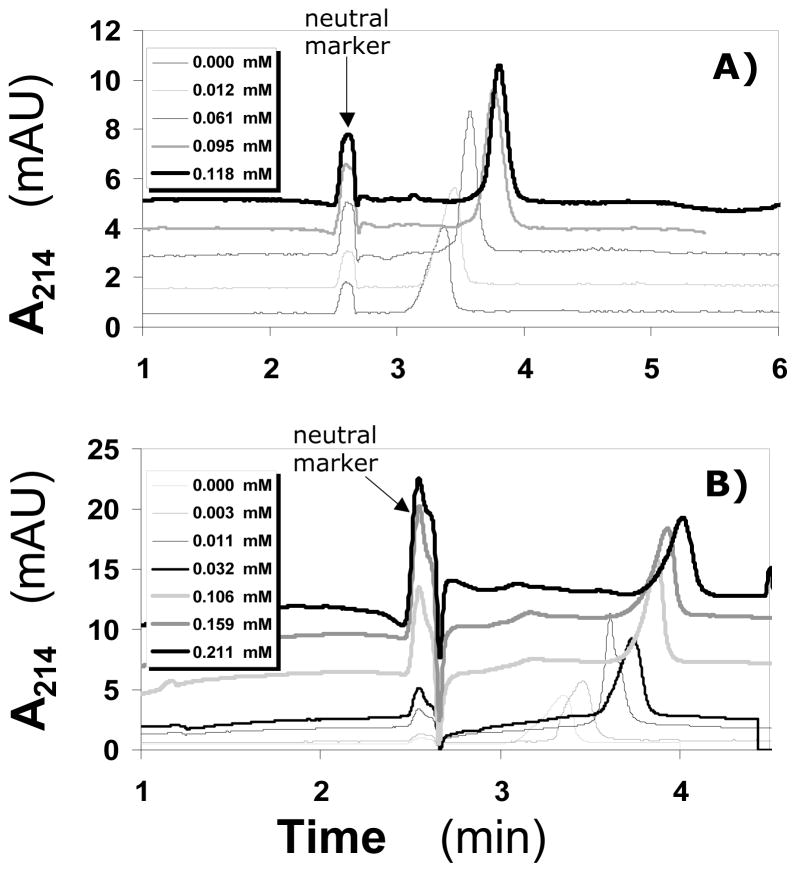
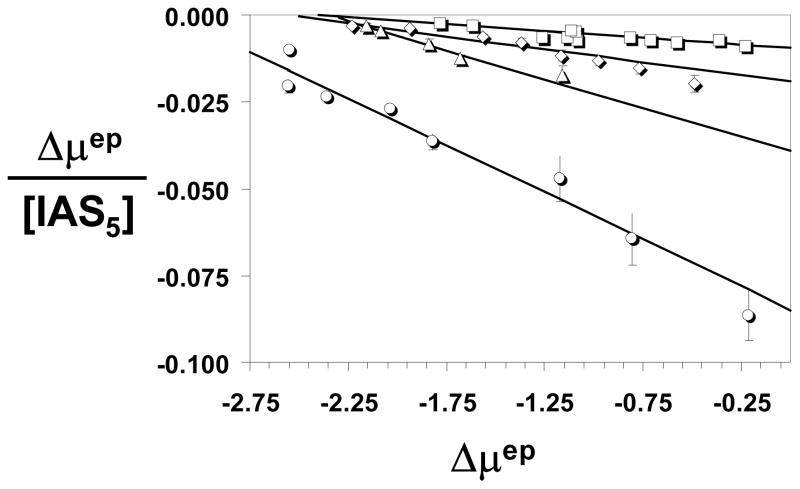
To test whether ACE can be used to study the small molecule -AT interaction, IAS5 was chosen. IAS5 was the only small aromatic molecule in the tetrahydroisoquinoline series (Fig. 1B) that induced a 30-fold increase in antithrombin inhibition of factor Xa.15 It also contained the most number of negatively charged groups, thereby potentially inducing a significant change in electrophoretic mobility of AT upon binding. Following a series of experiments to assess the concentration of AT necessary for optimal level signal to noise ratio and peak sharpness, 0.86 μM injection was chosen. A single AT peak was consistently observed suggesting rapid equilibrium between free and bound AT forms. Because IAS5 is negatively charged, the complex possesses an apparent higher charge-to-mass ratio than free AT resulting in the longer migration times with increasing concentrations of the ligand (Fig. 2A). The electropherograms indicate that IAS5 affected the mobility of AT in a concentration dependent manner. Analysis was performed in triplicate. The repeatability of migration times of the neutral marker and AT was 0.94% RSD and 1.03% RSD, respectively. Figure 3A shows the Scatchard plot of AT binding to IAS5 at pH 7.4. A linear Scatchard plot suggests that on average IAS5 binds to AT in one binding site, most probably in either PBS or EHBS (Fig. 1C). Equation 1 was used to fit the data to obtain a slope of −0.0233±0.0006 μM−1 corresponding to an equilibrium dissociation constant of 42.9±1.1 μM. This value corresponds well to a value of 37 μM measured using fluorescence.15
Figure 2.
Electropherograms of 0.86 μM AT in 20 mM sodium phosphate buffer, pH 7.4, containing different concentrations of synthetic sulfated molecules IAS5 (A) and IES5 (B). Naphthol was used as a neutral marker. Experimental conditions: Capillary: 31.2 cm total; 21 cm effective length; 75 μm I.D.; Applied voltage: 8 kV. Other conditions are described in ‘Materials and Methods’.
Figure 3.
Scatchard plots of the AT interaction with IAS5 (○), IES4 (◇), or IAS4 (□) (shown in Panel A) and with IES5 (○) (shown in Panel B). Electrophoretic mobilities (Δμep) in the presence of ligands (L) were measured as described in Materials and Methods. Solid line refers to the linear regressional fit to the data to give the binding affinity from the slope (= −1/KD). For IES5 (Panel B), appropriate data points were used to obtain two KD values. See text for a discussion on analysis.
Following the protocol set for IAS5, other synthetic derivatives were studied. IAS4 and IES4 both contain one less sulfate group in comparison to IAS5, while IES4 has a neutral ester group instead of the carboxylate group (Fig. 1B). The ACE profiles of IAS4 and IES4 interacting with AT were similar to IAS5 indicating these are also rapidly equilibrating systems with essentially similar characteristics. Analysis of the data using equation 1 gave slopes of −0.0212 ± 0.0004 and −0.0185 ± 0.0014 for IAS4 and IES4 implying KD values of 47.2 ± 0.8 μM and 54.1 ± 4.2 μM, respectively, for the complexes.
The interaction of IES5 with AT was studied using the same protocol as described above (Fig. 2B). IES5, which differs from IAS5 only in the absence of the carboxylate anion, displayed a biphasic Scatchard plot (Fig. 3B). This suggests that on average IES5 interacts with AT in two sites with sufficiently different binding affinities. Using Scatchard equation 1 to independently analyze the two arms of the biphasic profile gave slopes of −0.213±0.014 and −0.0330±0.0004 corresponding to KD values of 4.7 ± 0.6 and 30.3 ± 0.4 μM, respectively, in 20 mM sodium phosphate buffer, pH 7.4.
3.2 Nature of Interaction with AT
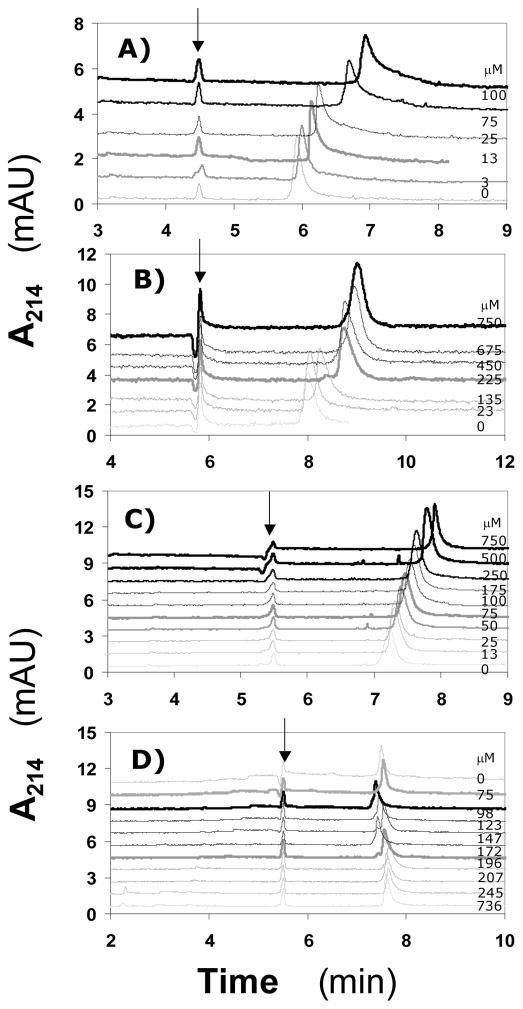
The aromatic as well as highly anionic character of these small synthetic ligands suggests that the interaction with AT could be driven by either hydrophobic or electrostatic forces. To determine the nature of interaction, the dissociation constant of IAS5 – AT interaction was measured (KD,OBS) as a function of NaCl concentration. Figure 4 shows the electropherograms of AT in the presence of varying concentrations of IAS5 in 20 mM sodium phosphate buffer, pH 7.4, containing 0, 10, 20 and 50 mM NaCl at 25 °C. The collection of electropherograms shows that as the concentration of NaCl increases, the magnitude of AT peak shift decreases, implying a decreasing affinity of interaction. This weakening affinity introduces significant difficulties in ACE measurements. In fact, the affinity of IAS5 for AT was not possible to measure reliably at concentrations higher than 50 mM NaCl. This could either be an intrinsic interaction limitation, e.g., too poor affinity, or it could be a limitation of CE, i.e., too much variation introduced by high current, temperature fluctuations, etc. Despite the difficulties associated with these measurements, the Scatchard plots of the interaction displayed good linearity in the range of concentrations studied (Fig. 5). The slopes of regression fits to the data were used to calculate KD values of 37, 60, 133 and 256 μM for IAS5 interacting with AT in the presence of 0, 10, 20 and 50 mM NaCl, respectively, at pH 7.4 (see Supplementary Information).
Figure 4.
Electropherograms of 3.4 μM AT in 20 mM sodium phosphate buffer, pH 7.4, containing 0 (Panel A), 10 (Panel B), 20 (Panel C) or 50 mM (Panel D) NaCl in the presence of varying concentrations of IAS5, which are displayed on the right hand side. Naphthol (arrow) was used as a neutral marker. Experimental conditions: Capillary: 40.2 cm total; 30 cm effective length; 50 μm I.D.; Applied voltage: 10 kV. Other conditions are described in ‘Materials and Methods’.
Figure 5.
Scatchard plots of the AT interaction with IAS5 in 20 mM phosphate buffer, pH 7.4, containing 0 (○), 10 (△), 20 (◇) and 50 mM NaCl (□) at 25 °C. Solid line represents linear fit to the data according to equation 1. See text for details.
The interaction of a polyanionic species, such as the highly sulfated ligands studied here, with a polycationic site, such as the heparin-binding site in AT, can be described as a protein – polyelectrolyte interaction, which has been extensively used earlier to describe heparin – AT interaction.7,8,28–30 According to this theory, the protein – polyelectrolyte interaction can be described by linear equation 3 in which the slope corresponds to the number of ion-pair interactions (Z) involved in binding, and the intercept corresponds to the binding energy due to non-ionic forces (KD,NON-IONIC). Thus, the contribution of ionic (ΔGOIONIC) and non-ionic (ΔGONON-IONIC) forces involved in the interaction can be obtained from the slope and intercept, respectively, of the linear plot. In this equation, Ψ is the fraction of monovalent counterion bound per activator ionic charge that is released on protein binding and has a value of 0.8.7,8
| (3) |
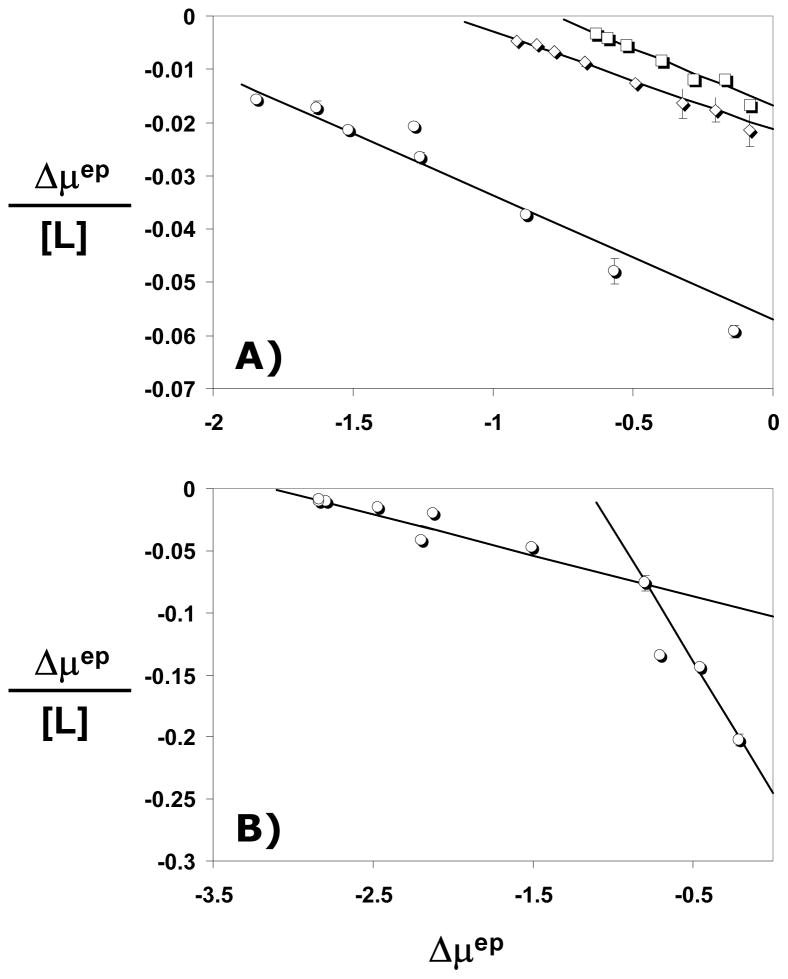
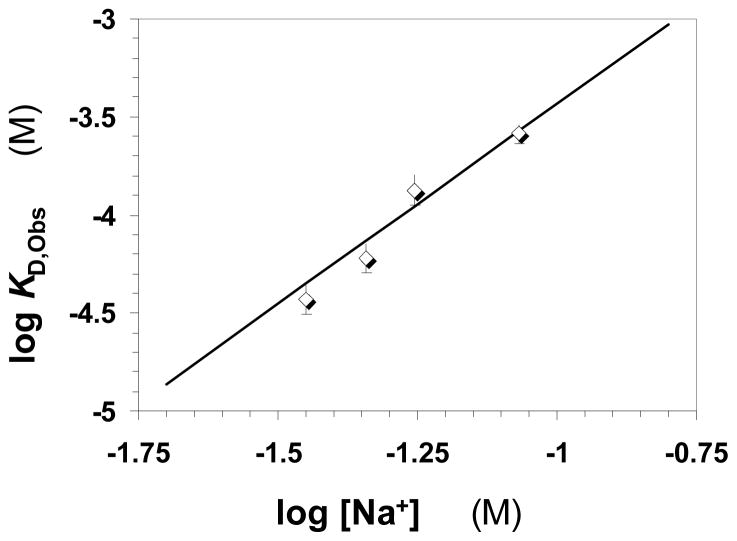
For IAS5, logKD increased linearly with log[Na+] at pH 7.4 (Fig. 6) suggesting an interaction profile similar to that of heparin or sulfated flavanoids with AT.7–9 Linear regression fit to the data gave a slope of 2.04±0.08, which implies that IAS5 makes 2.6±0.4 ion-pair interactions with AT at pH 7.4. Thus, approximately three negatively charged groups out of the six available are involved in interacting with AT. These three anions contribute 2.4±0.1 kcal/mol of free energy of binding due to ionic forces under near physiologic conditions, i.e., pH 7.4 containing 100 mM NaCl. The regression fit also gave an intercept of −1.39±0.09, which corresponds to a KD,NON-IONIC of 145±18 mM and a free energy of binding due to non-ionic forces of 1.9±0.2 kcal/mol at pH 7.4.
Figure 6.
Dependence of AT – IAS5 interaction affinity on the ionic strength of the medium at pH 7.4 and 25 °C. The binding affinities at various NaCl concentrations were measured as described in ‘Materials and Methods’ and shown in Figure 5. Solid line represents a linear regressional fit to the data using equation 3 to obtain the number of ionic interactions (Z) from the slope and the affinity due to non-ionic forces (KD, Non-ionic) from the intercept.
3.3 Site of Binding on AT
As highlighted in the introduction section, two contiguous, electropositive regions – PBS and EHBS (see Fig. 1C) – are known on AT.9,10 To determine where our non-saccharide activators bind AT, we studied the binding of IAS5 in the presence of two competitors. Pentasaccharide H5 (see Fig. 1A) was used for studying PBS competition, while CS, a rationally designed, sulfated flavanoid studied earlier (see Fig. 1B),9 was used for EHBS competition. H5 and CS have been shown to bind AT at pH 7.4 with 10 nM and 30 μM affinities, respectively.8,9 Thus, if IAS5 binds in PBS, the presence of H5 can be expected to decrease its affinity. Similarly, the presence of CS can be expected to weaken IAS5 affinity, if both sulfated ligands compete for the EHBS.
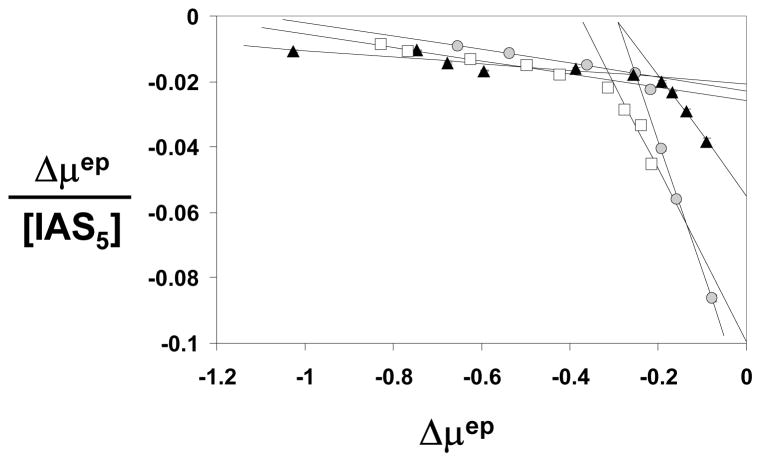
Competitive binding studies were performed in 20 mM sodium phosphate buffer, pH 7.4, containing 1, 5 and 10 nM H5 at 25 °C. The fixed concentrations of H5 induced a basal shift in the mobility of AT, which underwent further movement as a function of the increasing concentration of IAS5 (not shown). The highest H5 concentration possible to evaluate in this manner was 10 nM primarily because tight binding induced significant reduction in AT signal. Scatchard plots of the interaction at the three concentrations are shown in Figure 7. At all three H5 concentrations studied, IAS5 – AT interaction displays a biphasic profile. In contrast, the Scatchard plot of IAS5 alone was monophasic (see Figs. 3A and 5). Analysis of the biphasic data gave binding affinities of 2.5 and 48 μM at 1 nM H5, 5.4 and 99 μM at 5 nM H5, and 3.8 and 49 μM at 10 nM H5. In comparison, the AT affinity of IAS5 alone was measured to be ~40 μM under similar conditions.
Figure 7.
Competitive binding studies showing Scatchard plots of the IAS5 interacting with AT in the presence of 1 (○), 5 (△), and 10 nM (□) H5. Solid lines represent linear fit to the data using equation 1. See text for details.
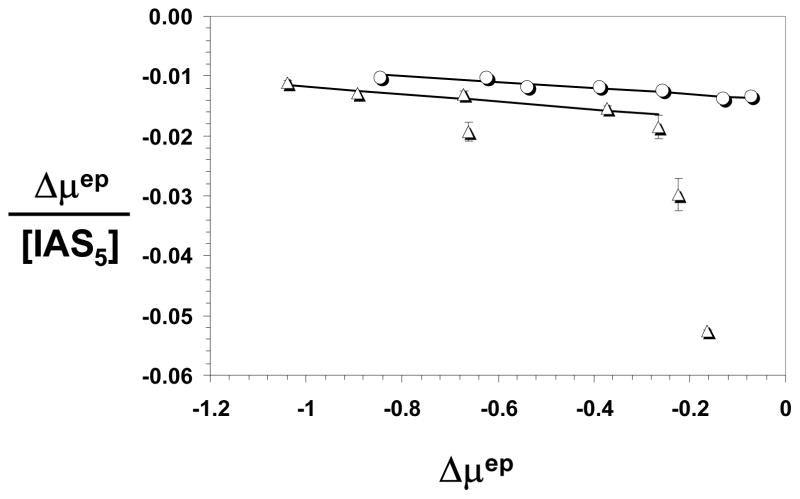
EHBS competitive studies were performed in the presence of 1 and 10 μM CS. In a manner similar to PBS competitive studies, the presence of CS induced a basal shift in the mobility of AT, which increased with IAS5. ACE profiles of AT – IAS5 interaction in the presence of higher concentrations of CS, e.g., 30 μM, led to considerable scatter. Figure 8 shows the Scatchard plots of the interaction at 1 and 10 μM CS. The Scatchard plot was found to be monophasic in the presence of 1 μM CS, while that in the presence of 10 μM CS was biphasic. Analysis of the data gave IAS5 binding affinities of 200±29 μM and 154±23 μM in the presence of 1 and 10 μM CS, respectively. The second binding affinity corresponding to the biphasic profile at 10 μM CS was measured to be 2.9±0.2 μM.
Figure 8.
Scatchard plots of theAT interaction with IAS5 in the presence of 1 (○) and 10 μM (△) CS. Solid lines represent linear fit to the data using equation 1. See text for details.
4 Discussion
Capillary electrophoresis has been previously used to assess the affinity of polyanionic polysaccharides for antithrombin. These polyanionic polysaccharides, e.g., heparins, readily alter the electrophoretic mobility of the glycoprotein due to the large number of negatively charged groups on the polymeric chain. Whether sulfated, small molecules would sufficiently modify the electrophoretic properties was an open question. We have found that ACE can be used to study AT – small molecule interaction if the binding affinity is in the range of nanomolar to ~200 μM. Thus, ACE is likely to be particularly suited for high-throughput screening of such small molecules because it is fast (< 10 minutes) and requires small amount of sample. Also, this protocol will be useful for small molecules that do not induce a significant change in spectroscopic properties, e.g., absorbance or fluorescence, of AT.
The study also uncovered interesting mechanistic aspects of small molecule – AT interaction. Despite the similarity in the structures of the tetrahydroisoquinoline-based molecules, it appears that their mode of interaction varies significantly. For example, whereas IAS5, IAS4 and IES4 exhibit a monophasic Scatchard plot with affinity in the range of 40 – 60 μM, IES5 exhibits a biphasic profile with ~5 and 30 μM affinities. The biphasic profile suggests the presence of an additional equilibrium for IES5 corresponding to a 5 μM affinity. The biphasic profile could arise from binding in both PBS and EHBS of AT. Alternatively, it could arise from an interaction with the same site – either PBS or EHBS – present in different forms of the inhibitor, i.e., the native form (AT) or the activated form (AT*), which exists in equilibrium (Fig. 9).8 We propose that the four small sulfated activators interact with EHBS present in the native form of AT with 30 – 60 μM affinity, while IES5 also interacts with EHBS present in the activated form of AT (AT*) with an affinity of 5 μM. This hypothesis is supported through competitive binding studies with pentasaccharide H5.
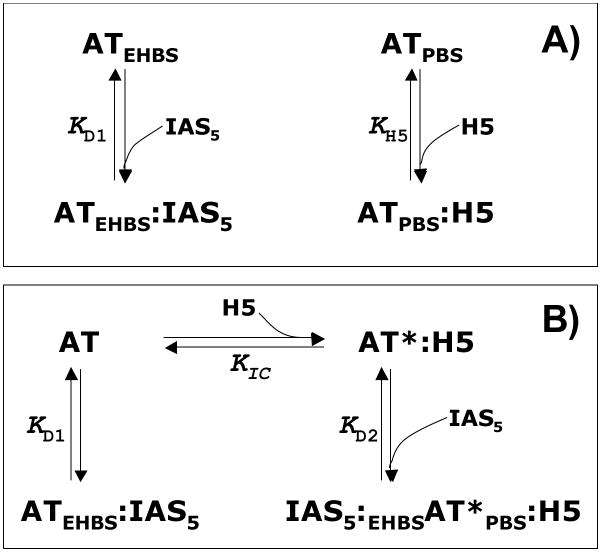
Figure 9.
Biochemical models of non-saccharide activator IAS5 binding to AT under various conditions. Panel A: Two sites – EHBS and PBS – are available in native AT (AT) for interaction with ligands, such as H5 and IAS5. H5 preferentially interacts with PBS with binding affinity KH5, while IAS5 prefers EHBS and binds with affinity KD1, which is ~40 μM. Panel B: The presence of H5 induces activation of native antithrombin (AT) to its activated state (AT*) generating an additional equilibrium involving AT*:H5 species. Thus, in the presence of H5, which binds in the PBS, IAS5 can bind in the EHBS of either native (AT) or activated antithrombin (AT*) resulting in the formation of ATEHBS:IAS5 or IAS5:EHBSAT*PBS:H5 complexes, respectively. The affinity of ATEHBS:IAS5 remains unaltered (~40 μM), while the affinity of IAS5:EHBSAT* was measured to be 2 – 5 μM in 20 mM sodium phosphate buffer, pH 7.4 at 25 °C. See Discussion section for details.
The AT – H5 interaction is extremely well characterized in terms of both structure and biochemistry.7,8,28–34 It is well-established that H5 selectively engages the inhibitor in the PBS. Thus, competitive binding studies with H5 were performed to test whether IAS5 binds in the PBS. Interestingly, instead of an expected monophasic profile, the Scatchard plot for IAS5 – AT interaction in the presence of H5 was biphasic. This indicated the operation of two equilibria with sufficiently distinct binding constants. Further, the two KD values remained essentially invariant as the concentration of H5 increased from 1 to 10 nM. Of these, the weaker interaction (~50 – 100 μM) was similar in magnitude to that measured in the absence of H5 (40 μM). Thus, the presence of H5 introduces a new, stronger IAS5 – AT interaction with 2 – 5 μM affinity. Previous biochemical studies have shown that interaction of H5 in the PBS of AT activates the inhibitor to the activated form (AT*).7,8 This activation induces conformational changes in the binding site including the formation of helix P and ordering of residues in the EHBS.31–34 Thus, our observations suggest that engagement of the PBS by H5 induces better recognition of the EHBS by IAS5 (see Fig. 9B), which is reflected in the biphasic Scatchard plot.
To confirm that IAS5 preferentially recognizes EHBS, competitive binding studies were performed in the presence of CS, a small, aromatic flavanoid known to bind in the EHBS.9 Presence of 1 μM CS induced a weakening in AT of IAS5 affinity from 37 μM to 200 μM, which resembles competitive binding. Yet, the magnitude of the affinity change (~5-fold) is more than that predicted on the basis of Dixon-Webb relationship (KD, APP = KD{1+[I]/KI}), which describes an ideal competitive inhibition behavior.8,9 Another evidence for movement away from strictly competitive inhibition is described by 10 μM CS, which introduces a biphasic profile with 154 and 2.9 μM KD values. One reason for this biphasic profile could be simultaneous occupation of PBS and EHBS by the two activators. Thus, the presence of CS does introduce competition for binding in EHBS, however, it appears that the multiple anionic groups present in these molecules may seek out the PBS under forcing condition, e.g., high concentration.
A final question related to understanding the forces involved in the recognition of these highly sulfated molecules, which is extremely important for advancing the rational design. Ionic strength dependence of binding affinity indicated that IAS5 formed 2.6 ion-pair interactions with AT. This implies that nearly 3 negative charges out of the 6 available in IAS5 are involved in binding. Our previous studies with CS indicated the formation of 2 ion-pair interactions in EHBS,9 while full-length heparin is known to form one interaction in the EHBS.7,8 In terms of energy, these interactions correspond to an ionic binding energy of 2.4 kcal/mol under physiological conditions. In contrast, the non-ionic contribution was found to be 1.9 kcal/mol. Thus, nearly 44% of binding energy arises from non-ionic forces, which is similar to that observed in the case of H5 and CS interacting with AT.7–9 We speculate that this non-ionic binding energy arises from hydrogen bond-type interactions involved in sulfate and carboxylate groups recognizing electropositive residues in the EHBS, as noted for H5 – AT.
Overall, our work exploits the power of ACE in understanding antithrombin interactions with small highly charged molecules at a molecular level. Mechanistically, our small molecules primarily engage the EHBS, which explains their weak potency in activating the inhibitor. ACE was found to be particularly suited for this work. More importantly, it is expected to be very useful for high-throughput screening of small molecules toward the discovery of new anticoagulants.
Supplementary Material
Table.
Interaction of IAS5 with Antithrombin in the presence of varying concentrations of NaCl at pH 7.4 and 25 °C.@
| [NaCl] | Slope | Intercept | KD@ |
|---|---|---|---|
| (mM) | (μM−1) | (cm2 μM−1min−1kV−1) | (μM) |
| 0 | −0.0270 ± 0.0040$ | −0.0850 ± 0.0040 | 37 ± 6 |
| 10 | −0.0167 ± 0.0024 | −0.0390 ± 0.0010 | 60 ± 8 |
| 20 | −0.0075 ± 0.0006 | −0.0190 ± 0.0030 | 133 ± 10 |
| 50 | −0.0039 ± 0.0002 | −0.0094 ± 0.0007 | 256 ± 12 |
The binding affinities at 0, 10, 20 and 50 mM NaCl were measured as described in ‘Materials and Methods’ and shown in Figure 5. Scatchard analysis was performed to obtain the slope and intercept from which KD values were calculated.
Errors represent ±2 S. E.
Acknowledgments
The authors thank Professor Steven Olson of University of Illinois – Chicago for the generous gift of heparin pentasaccharide. This work was supported in part by the National Heart, Lung and Blood Institute (RO1 HL069975), the American Heart Association National Center (EIA 0640053N), the Mizutani Foundation for Glycoscience, and the A. D. Williams Foundation (6-48708 and 6-46604).
Abbreviations
- ACE
affinity capillary electrophoresis
- AT
antithrombin
- CE
capillary electrophoresis
- EHBS
extended heparin-binding site
- HBS
heparin-binding site
- PBS
pentasaccharide-binding site
References
- 1.Bjork I, Olson ST. Adv Exp Med Biol. 1997;425:17–33. [PubMed] [Google Scholar]
- 2.Olson ST, Swanson R, Raub-Segall E, Bedsted T, Sadri M, Petitou M, Herault JP, Herbert JM, Bjork I. Thromb Haemost. 2004;92:929–939. doi: 10.1160/TH04-06-0384. [DOI] [PubMed] [Google Scholar]
- 3.Desai UR. Med Res Rev. 2004;24:151–158. doi: 10.1002/med.10058. [DOI] [PubMed] [Google Scholar]
- 4.Hirsh J, Anand SS, Halperin JL, Fuster V. Circulation. 2001;103:2994–3018. doi: 10.1161/01.cir.103.24.2994. [DOI] [PubMed] [Google Scholar]
- 5.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser S, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan S. Nat Biotech. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishimoto TK, Viswanathan K, Ganguly T, Elankumaran S, Smith S, Pelzer K, Lansing JC, Sriranganathan N, Zhao G, Galcheva-Gargova Z, Al-Hakim A, Bailey GS, Fraser B, Roy R, Rogers-Cotrone T, Buhse L, Whary M, Fox J, Nasr M, Dal Pan GJ, Shriver Z, Langer RS, Venkataraman G, Austen KF, Woodcock J, Sasisekharan R. N Engl J Med. 2008;358:2457–2467. doi: 10.1056/NEJMoa0803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson ST, Bjork I, Sheffer R, Craig PA, Shore JD, Choay J. J Biol Chem. 1992;267:12528–12538. [PubMed] [Google Scholar]
- 8.Desai UR, Petitou M, Bjork I, Olson ST. J Biol Chem. 1998;273:7478–7487. doi: 10.1074/jbc.273.13.7478. [DOI] [PubMed] [Google Scholar]
- 9.Gunnarsson GT, Desai UR. J Med Chem. 2002;45:4460–4470. doi: 10.1021/jm020132y. [DOI] [PubMed] [Google Scholar]
- 10.Arocas V, Turk B, Bock SC, Olson ST, Bjork I. Biochemistry. 2000;39:8512–8518. doi: 10.1021/bi9928243. [DOI] [PubMed] [Google Scholar]
- 11.Gunnarsson GT, Desai UR. J Med Chem. 2002;45:1233–1243. doi: 10.1021/jm020012q. [DOI] [PubMed] [Google Scholar]
- 12.Gunnarsson GT, Riaz M, Adams J, Desai UR. Bioorg Med Chem. 2005;13:1783–1789. doi: 10.1016/j.bmc.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 13.Gunnarsson GT, Desai UR. Bioorg Med Chem Lett. 2003;13:579–583. doi: 10.1016/s0960-894x(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 14.Raghuraman A, Riaz M, Hindle M, Desai UR. Tetrahedron Lett. 2007;48:6754–6758. doi: 10.1016/j.tetlet.2007.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghuraman A, Liang A, Krishnasamy C, Lauck T, Gunnarsson GT, Desai UR. Eur J Med Chem. 2008 doi: 10.1016/j.ejmech.2008.09.042. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyler Cross R, Sobel M, Marques D, Harris RB. Prot Sci. 1994;3:620–627. doi: 10.1002/pro.5560030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl U, Thunberg L, Backstrom G, Riesenfeld J, Nordling K, Bjork I. J Biol Chem. 1984;259:12368–12376. [PubMed] [Google Scholar]
- 18.Pijler G, Backstrom G, Lindahl U. J Biol Chem. 1987;262:5036–5043. [PubMed] [Google Scholar]
- 19.Osmond Ronald IW, Kett Warren C, Skett Spencer E, Coombe Deirdre R. Anal Biochem. 2002;310:199–207. doi: 10.1016/s0003-2697(02)00396-2. [DOI] [PubMed] [Google Scholar]
- 20.Mille S, Watton J, Barrowcliffe TW, Mani JC, Lane DA. J Biol Chem. 1994;269:29435–29443. [PubMed] [Google Scholar]
- 21.Gunnarsson K, Valtcheva L, Hjerten S. Glycoconj J. 1997;14:859–862. doi: 10.1023/a:1018594122670. [DOI] [PubMed] [Google Scholar]
- 22.Le Saux T, Varenne A, Perreau F, Siret L, Duteil S, Duhau L, Gareil P. J Chromatogr A. 2006;1132:289–296. doi: 10.1016/j.chroma.2006.07.092. [DOI] [PubMed] [Google Scholar]
- 23.Seyrek E, Dubin PL, Henriksen J. Biopolymers. 2007;86:249–259. doi: 10.1002/bip.20731. [DOI] [PubMed] [Google Scholar]
- 24.Varenne A, Gareil P, Colliec-Jouault S, Daniel R. Anal Biochem. 2003;315:152–159. doi: 10.1016/s0003-2697(02)00687-5. [DOI] [PubMed] [Google Scholar]
- 25.Wu XJ, Linhardt RJ. Electrophoresis. 1998;19:2650–2653. doi: 10.1002/elps.1150191514. [DOI] [PubMed] [Google Scholar]
- 26.Colton IJ, Carbeck JD, Rao J, Whitesides GM. Electrophoresis. 1998;19:367–382. doi: 10.1002/elps.1150190303. [DOI] [PubMed] [Google Scholar]
- 27.Scatchard G. Ann N Y Acad Sci. 1949;51:660–672. [Google Scholar]
- 28.Desai UR, Petitou M, Bjork I, Olson ST. Biochemistry. 1998;37:13033–13041. doi: 10.1021/bi981426h. [DOI] [PubMed] [Google Scholar]
- 29.Olson ST, Halvorson HR, Bjork I. J Biol Chem. 1991;260:6342–6352. [PubMed] [Google Scholar]
- 30.Olson ST, Bjork I. J Biol Chem. 1991;260:6353–6364. [PubMed] [Google Scholar]
- 31.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. Proc Natl Acad Sci. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson DJ, Huntington JA. Biochemistry. 2003;42:8712–8719. doi: 10.1021/bi034524y. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Johnson DJ, Esmon CT, Huntington JA. Nat Struct Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 34.Dementiev A, Petitou M, Herbert JM, Gettins PG. Nat Struct Mol Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.