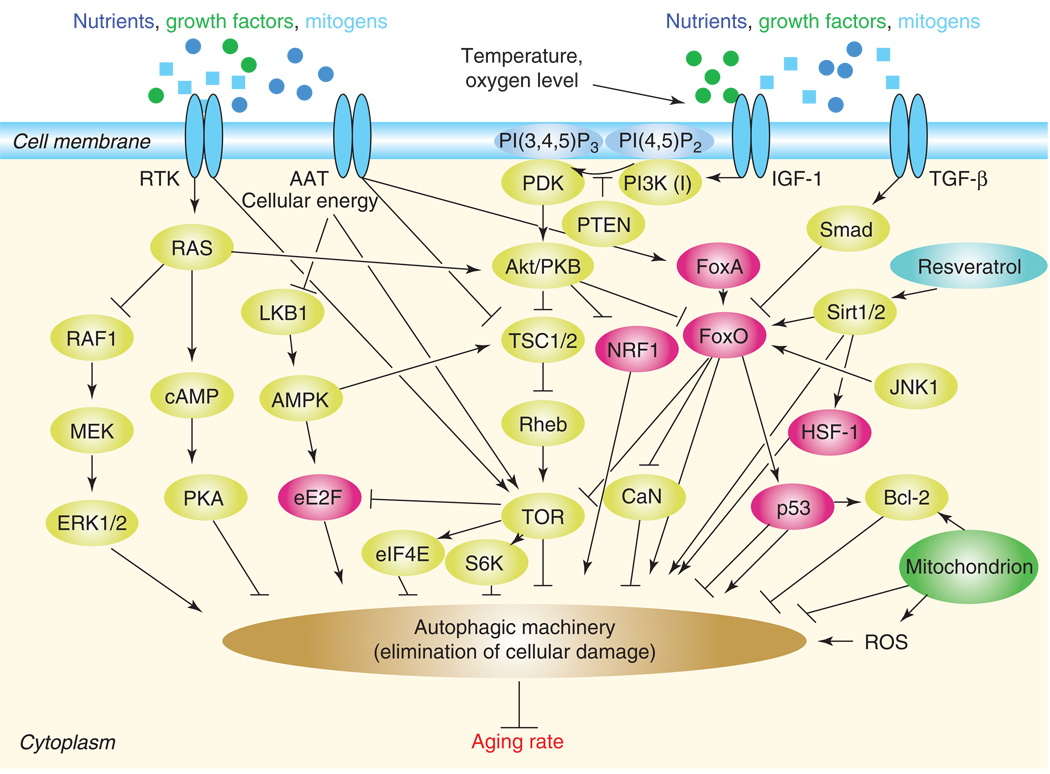
Figure 1. Model for the signaling network of aging.
The overall rate at which cellular damage accumulates is regulated by multiple pathways (yellow components) and transcription factors (red) that adjust mechanisms for maintenance and repair. These transcription factors influence autophagy through modulating the activity of autophagy-related genes in the nucleus (for simplicity, the nucleus is not shown). Autophagy (brown) is the main cellular degradation process for eliminating or recycling damaged cellular constituents (the proteasomal system also plays an important role in eliminating damage and thereby in aging, but it is not shown here). Longevity pathways, which respond to diverse environmental cues such as nutrient availability, temperature and oxygen level, converge on the autophagic pathway to control aging. The chemical substance resveratrol is indicated by blue coloring, the mitochondrion is green. This figure illustrates the mammalian counterparts of signaling and metabolic components involved in aging. Several interrelationships among these components have only been demonstrated thus far in model organisms. Arrows (activations) and bars (inhibitions) indicate direct interactions that have been experimentally confirmed. However, not all the indirect interactions have as yet necessarily been demonstrated (e.g., it is unknown whether Sirt1/2 regulate eE2F activity). RTK: receptor tyrosine kinase; AAT: amino acid transporters; IGF-1: insulin-like growth factor-1 receptor; PI(4,5)P2; phosphatidylinositol (4,5)-bisphosphate; PI(3,4,5)P3: phosphatidylinositol (3,4,5)-trisphosphate; PI3K (I): class I phosphatidylinositol 3-kinase; PI3K (III): class III phosphatidylinositol 3-kinase; TGF-β: transforming growth factor-beta receptor; ROS: reactive oxygen species; Bcl-2: B cell lymphoma-2; JNK1: c-Jun N-terminal kinase-1; HSF-1: heat shock response transcription factor-1; NFR1: isoform 1 of nuclear factor erythroid 2-related factor 2; FoxO: Forkhead-family transcription factor; FoxA: FoxA transcription factor; Sirt1/2: NAD-dependent histone deacetylase-1/2; Smad: SMAD family member; PTEN: phosphatase and tensin homolog; Akt/PKB: AKT8 virus protooncogene/protein kinase B; PDK: 3-phosphoinositide-dependent protein kinase; CaN: calcineurin; TSC1/2: tuberous sclerosis complex-1/2; Rheb: Ras homologue enriched in brain; TOR: target of rapamycin; S6K: S6 kinase; eIFE4: eukaryotic translation initiation factor; eE2F: eukaryotic transcription factor; AMPK: AMP-activated protein kinase; LKB1: serine/threonine protein kinase; PKA: protein kinase A; ERK1/2: extracellular signal-related tyrosine kinase.