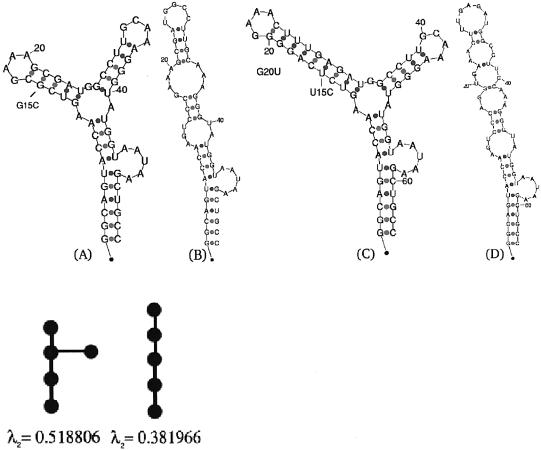
Figure 1.
The secondary structure of the P5abc subdomain of the T.thermophila group I intron ribozyme. (A) The wild-type folded structure, along with its tree graph representation and the corresponding second eigenvalue below it. The tree graph possesses five vertices, since a ‘pseudo-loop’ representing the 5′-3′ is uniformly accounted for as a node, in addition to the loops, bulges and hairpins. An artificial structure without 4 bp near the L5b GAAA tetraloop has been shown by NMR experiments (21) to yield the same secondary structure predicted by mfold without the addition of magnesium ions. (B) Mutated structure; predicted mutation G15C with respect to the wild-type structure in (A), along with its tree graph representation and the corresponding second eigenvalue below it. (C) The predicted wild-type folded structure of the natural P5abc subdomain, with the 4 bp included in the P5b stem, as in Cate et al. (22). (D) Mutated structure; predicted mutation U15C and G20U with respect to the wild-type structure in (C). A two point mutation is required to disrupt the L5b GAAA tetraloop in case the 4 bp are not removed, when using the deleterious mutation prediction method described in the text.