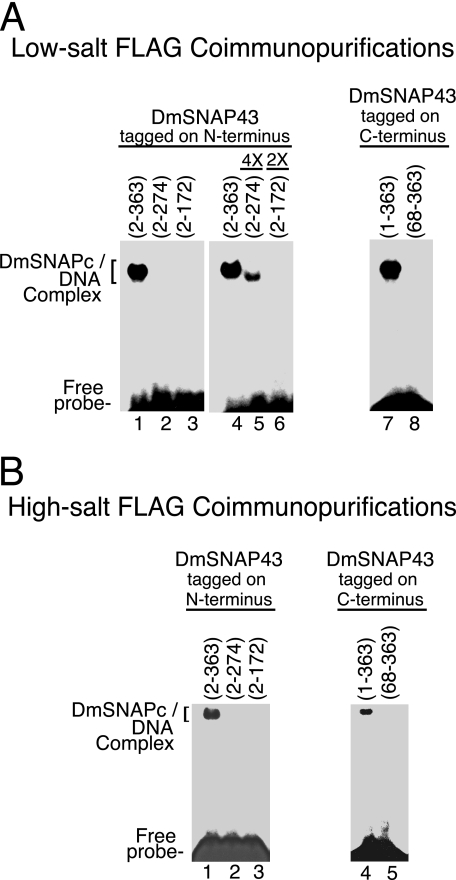
FIGURE 7.
Domains of DmSNAP43 required for sequence-specific DNA binding by DmSNAPc. A, DmSNAP complexes FLAG affinity purified under low-salt conditions (Fig. 6C) were used for DNA mobility shift analysis with a DNA probe containing a U1 PSEA sequence. Complexes containing DmSNAP43 constructs tagged at the N terminus (C-terminal truncations) are shown in lanes 1–6, and constructs tagged at the C terminus (N-terminal truncations) are shown in lanes 7 and 8. Lanes 1–4 and 7 and 8 were carried out with complexes containing a normalized amount of DmSNAP43 as determined in Fig. 6C, whereas lanes 5 and 6 contained 4 or 2 times the normalized amount of truncated protein as indicated above each lane. (The maximum amount of protein that could be used was limited by the volume of the reaction and the relative concentration of each sample.) B, similar to A except DmSNAP complexes were affinity purified under high-salt conditions and protein amounts were normalized based upon the immunoblots shown in Fig. 6D.