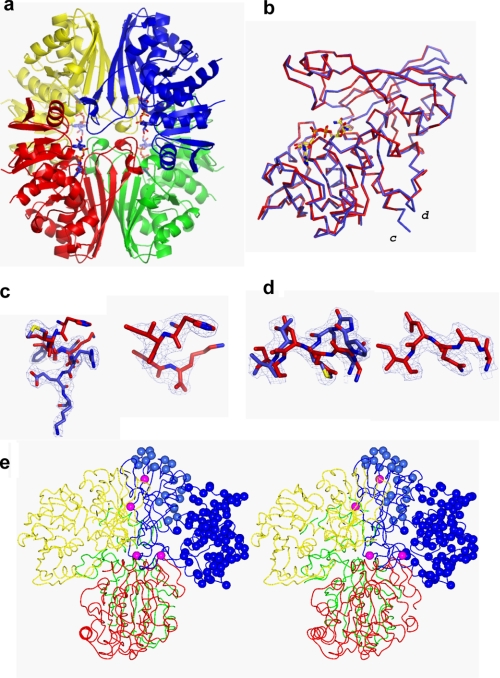
FIGURE 4.
Structure of His-GAPDS E. coli GAPDH heterotetramer. a, ribbon diagram of the His-GAPDS-E. coli GAPDH tetramer. The tetramer is formed through the association of a pair of dimers. Chain A (red) and chain C (green) form an E. coli GAPDH dimer, and chain B (yellow, E. coli GAPDH) and chain D (blue, His-GAPDS) form a heterodimer. NAD+ is shown in sticks representation. b, chain A, E. coli GAPDH (red) overlaid on chain D, GAPDS (blue). c, C-terminal extension in D subunit. The final refined model for the sequence 328YMFSREK334 His-GAPDS is shown in blue bonds representation, with final weighted electron density shown at 1σ. The final refined model for 327HISK330 E. coli GAPDH (red) is superimposed. On the right-hand side, the final refined coordinates for the C terminus of E. coli GAPDH, 327HISK330, are shown in red bonds representation with final weighted electron density shown at 1σ. d, final refined model for the sequence 138NPGSMTV146 His-GAPDS is shown in blue bonds representation with final weighted electron density shown at 1σ, and the final refined model for the corresponding sequence in E. coli from subunit A, 139AGQDI143 E. coli GAPDH, is superimposed in red bonds representation. On the right-hand side, the same region 139AGQDI143 E. coli GAPDH (red) is shown with its own final weighted electron density shown at 1σ. e, trace of tetramer with residue positions that differ in sequence between E. coli GAPDH (A, red; B, yellow; C, green) and GAPDS (blue) shown as blue spheres in the D subunit. Residues that lie at the interfaces between subunits are colored magenta.