FIGURE 6.
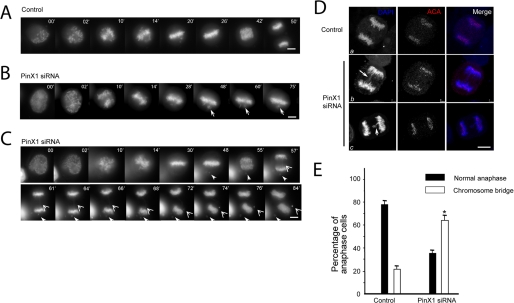
PinX1 governs faithful chromosome segregation. A, real time imaging of chromosome movements in HeLa cells transfected with scramble siRNA oligonucleotides. Chromosomes were marked by GFP-H2B. Bar, 10 μm. B and C, repression of PinX1 by siRNA treatment resulted in chromosome instability phenotype. Chromosome segregation errors are apparent in the absence of PinX1; closed arrows indicate lagging chromosomes during alignment; open arrows indicate chromosome bridges in anaphase. Bars, 10 μm. D, depletion of PinX1 impairs spindle checkpoint by promoting a premature anaphase (panels b and c). HeLa cells were transfected with PinX1 siRNA oligonucleotide and control oligonucleotide for 48 h followed by fixation and indirect immunofluorescence staining. This set of optical images was collected from an anaphase HeLa cell doubly stained for human ACA (red), DAPI (DNA, blue), and their merged images. As shown in panels b and c, PinX1-depleted cell entered in anaphase with a chromatin bridge (panels b and c, arrow). Most interestingly, ACA staining (panel c, arrowhead) verifies that this pair of chromatids failed to separate as the cell entered anaphase, leading to occurrence of aneuploidy. Bars, 10 μm. E, depletion of PinX1 by siRNA effects mitotic defects in chromosome segregation. HeLa cells were transfected with PinX1 siRNA oligonucleotide and control oligonucleotide for 48 h followed by fixation and DNA staining. Cells were then examined under a fluorescence microscopy to score the sister chromatid bridge phenotype. An average of 200 cells from three separate experiments was counted; *, p < 0.01 compared with that of the control.