FIGURE 4.
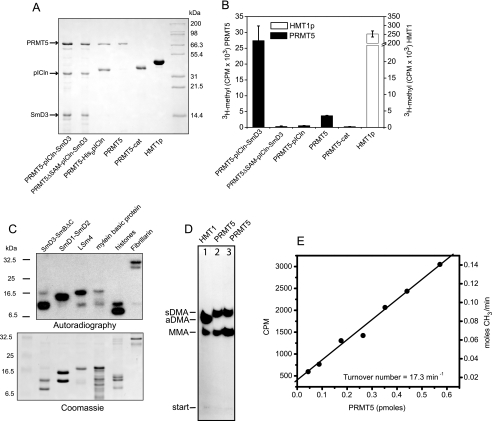
Active PRMT5 produced by co-expression with pICln. A, SDS-PAGE of PRMT5 complexes produced by co-expression with pICln in E. coli. PRMT5ΔSAM is a catalytically defective PRMT5 mutant. PRMT5 alone was obtained by separation of the purified PRMT5·pICln complex. The PRMT5 catalytic domain (296–637) and yeast HMT1 were expressed independently. B, arginine methylation activity of PRMT5 complexes. Reactions containing 20 μm SmD3/SmBΔC substrate and 40 μm [3H]SAM were separated after 1 h, and the amount of substrate methylation was determined by scintillation counting. Error bars represent the S.D. based on three replicates. C, methylation of RG-containing substrates by PRMT5. Reactions containing 1 μm PRMT5, 40 μm [3H]SAM (1 μCi/reaction) and the indicated substrates were incubated for 2 h followed by SDS-PAGE and autoradiography. D, recombinant PRMT5 produces monomethylarginine (MMA) and symmetric dimethylarginine (sDMA) but not asymmetric dimethylarginine (aDMA). Methylation reactions containing 5 μm HMT1 + 5 μm pICln·SmD3 (lane 1), 5 μm PRMT5·pICln·SmD3 complex (lane 2), or 5 μm PRMT5·pICln·SmD3 + 5 μm pICln·SmD3 (lane 3) were incubated with 80 μm [3H]SAM (20 μCi/reaction), and the resulting modified arginine residues were analyzed by thin layer chromatography and autoradiography as described (8). E, PRMT5 specific activity was determined from methylation reactions containing varying amounts of PRMT5 and saturating levels (20 μm) of pICln·SmD3 and [3H]SAM (80 μm SAM, 1 μCi/ reaction).