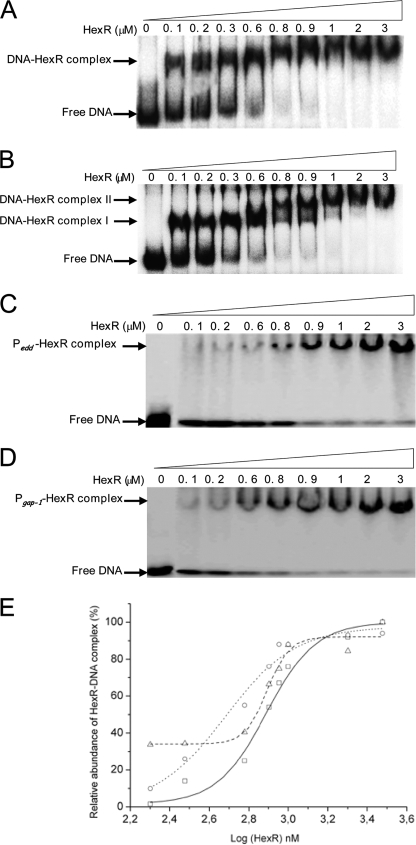
FIGURE 5.
Electrophoresis mobility shift assay of the promoter region of P zwf and Pedd/gap-1 by purified HexR. A, the DNA binding assays was done using purified HexR with 2 nm concentrations of the product of PCR (280 bp) of Pzwf end-labeled with 32P and incubated with the concentration of purified HexR indicated at the top of each lane. The position of the free DNA is indicated. B, as in A, but a 340-bp Pedd/gap-1 fragment was used. Complex I and Complex II are indicated. C and D, as in A, but a 280-bp DNA fragment containing the HexR binding site at Pgap-1 (D) or Pedd (C) were used. E, the densitometric analysis of the EMSA gels. Squares, Pzwf promoter fragments; circles, Pedd promoter fragments; triangles, and Pgap-1 promoter fragments.