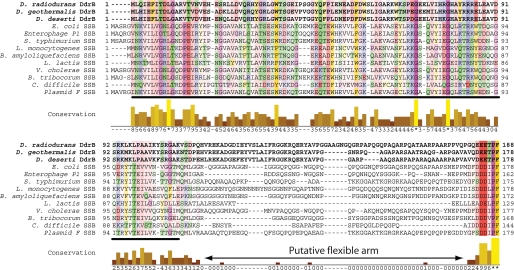
FIGURE 3.
An alignment of DdrB and bacterial SSB sequences using Clustal-W. DdrBs and SSBs exhibit very limited homology in the OB-fold domain (underlined) but share an analogous C-terminal motif ([DE](3)-X-PF>, where X is Thr in DdrB and a hydrophobic residue in SSB proteins) used by SSBs to interact with partners. The flexible arm between the OB-fold domain and the conserved C-terminal motif can be distinguished as the region of limited sequence similarity. DdrB from the three sequenced Deinococcus species were compared with a panel of bacterial SBB proteins: NP_418483 from E. coli, YP_006485 from Enterobacteria phage P1, BAB91629 from Salmonella typhimurium, CAC98260 from Listeria monocytogenes, ABS76124 from Bacillus amyloliquefaciens FZB42, AAL02359 from Lactococcus lactis subsp. cremoris, AAC72352 from Vibrio cholerae, CAK01796 from Bartonella tribocorum CIP 105476, YP_001090187 from Clostridium difficile 630, and BAA97930 from Plasmid F.