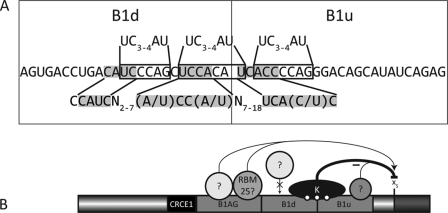
FIGURE 7.
A, putative binding sites for hnRNP K in the B1d and B1u elements. The three sites resembling the high affinity binding sites are boxed and compared with those identified by SELEX (48), shown above the RNA sequence for the B1 element. Below is the binding site defined using yeast three-hybrid screens, compared with the shaded binding site found on the Bcl-x pre-mRNA. B, hnRNP K binding to B1 may compete with the binding of an unknown enhancer protein, thus inhibiting splicing to the Bcl-xS site. Deletion of the enhancer element B1AG augments the use of the Bcl-xL splice site. When the other enhancer element, B1d, is removed, the inhibition by hnRNP K on the Bcl-xS 5′ splice site would be slightly diminished, perhaps by the stronger binding of a protein to B1AG or reduced binding affinity of hnRNP K to the pre-mRNA. Removal of the silencer B1u would abrogate hnRNP K binding leading to a strong increase in the production of the Bcl-xS isoform, as does a knockdown of hnRNP K by RNA interference.